Ex 2: Recruitment linked to an environmental covariate (Cold Pool Index)
Source:vignettes/ex2_CPI_recruitment.Rmd
ex2_CPI_recruitment.RmdIn this vignette we walk through an example using the
wham (WHAM = Woods Hole Assessment Model) package to run a
state-space age-structured stock assessment model. WHAM is a
generalization of code written for Miller et al. (2016)
and Xu et
al. (2018), and in this example we apply WHAM to the same stock,
Southern New England / Mid-Atlantic Yellowtail Flounder.
This is the 2nd wham example, which builds off model
m4 from example 1 (full state-space model, numbers at all
ages are random effects, logistic normal age-compositions). We assume
you already have wham installed. If not, see the Introduction. The simpler
1st example, without environmental effects, is available as a R
script and vignette.
In example 2, we demonstrate how to specify and run WHAM with varying
recruitment models (random, Bev-Holt, Ricker)
environmental covariate (Cold Pool Index, CPI) process models (random walk, AR1), and
how the CPI affects recruitment (controlling or limiting)
As in example
1, we check that each model converges
(check_convergence()), plot diagnostics, results, and
reference points (plot_wham_output()), and compare models
using AIC and Mohn’s rho (compare_wham_models()).
1. Prepare wham
Open R and load the wham package:
For a clean, runnable .R script, look at
ex2_CPI_recruitment.R in the example_scripts
folder of the wham package install:
wham.dir <- find.package("wham")
file.path(wham.dir, "example_scripts")You can run this entire example script with:
write.dir <- "choose/where/to/save/output" # otherwise will be saved in working directory
source(file.path(wham.dir, "example_scripts", "ex2_CPI_recruitment.R"))Let’s create a directory for this analysis:
# choose a location to save output, otherwise will be saved in working directory
write.dir <- "choose/where/to/save/output"
dir.create(write.dir)
setwd(write.dir)WHAM was built by modifying the ADMB-based ASAP model code (Legault and
Restrepo 1999), and is designed to take an ASAP3 .dat file as input.
We generally assume in wham that you have an existing ASAP3
.dat file. If you are not familiar with ASAP3 input files, see the ASAP
documentation
and code. For this
vignette, an example ASAP3 input file is provided,
ex2_SNEMAYT.dat. We will also need a data file with an
environmental covariate, the Cold Pool Index, CPI.csv.
Copy ex2_SNEMAYT.dat and CPI.csv to our
analysis directory:
wham.dir <- find.package("wham")
file.copy(from=file.path(wham.dir,"extdata","ex2_SNEMAYT.dat"), to=write.dir, overwrite=FALSE)
file.copy(from=file.path(wham.dir,"extdata","CPI.csv"), to=write.dir, overwrite=FALSE)Confirm you are in the correct directory and it has the required data files:
list.files()
#> [1] "age_comp_likelihood_test_values.rds" "BASE_3"
#> [3] "CPI.csv" "ex1_SNEMAYT.dat"
#> [5] "ex1_test_results.rds" "ex11_tests.rds"
#> [7] "ex2_SNEMAYT.dat" "ex2_test_results.rds"
#> [9] "ex4_test_results.rds" "ex5_summerflounder.dat"
#> [11] "ex5_test_results.rds" "ex6_test_results.rds"
#> [13] "ex7_SNEMAYT.dat" "GSI.csv"Read the ASAP3 .dat file into R and convert to input list for wham:
asap3 <- read_asap3_dat("ex2_SNEMAYT.dat")Load the environmental covariate (Cold Pool Index, CPI) data into R:
env.dat <- read.csv("CPI.csv", header=T)We generally abbreviate ‘environmental covariate’ as
ecov in the code. In this example, the ecov
data file has columns for observations (CPI), standard
error (CPI_sigma), and year (Year).
Observations and year are always required. Standard error can be treated
as fixed/data with yearly values (as here) or one overall value shared
among years. It can also be estimated as a parameter(s), likewise either
as yearly values or one overall value.
head(env.dat)
#> Year CPI CPI_sigma
#> 1 1973 0.5988 0.2838
#> 2 1974 -0.1760 0.2465
#> 3 1975 -1.1887 0.2539
#> 4 1976 -0.7938 0.2634
#> 5 1977 -0.6771 0.1576
#> 6 1978 -1.5195 0.20452. Specify models
Now we specify how the 7 models treat recruitment, the CPI process, and how the CPI affects recruitment:
df.mods <- data.frame(Recruitment = c(2,2,3,3,3,3,4),
ecov_process = c(rep("rw",4),rep("ar1",3)),
ecov_how = c(0,1,0,2,2,1,1), stringsAsFactors=FALSE)
n.mods <- dim(df.mods)[1]
df.mods$Model <- paste0("m",1:n.mods)
df.mods <- dplyr::select(df.mods, Model, tidyselect::everything()) # moves Model to first colLook at the model table
df.mods
#> Model Recruitment ecov_process ecov_how
#> 1 m1 2 rw 0
#> 2 m2 2 rw 1
#> 3 m3 3 rw 0
#> 4 m4 3 rw 2
#> 5 m5 3 ar1 2
#> 6 m6 3 ar1 1
#> 7 m7 4 ar1 1We specify the options for modeling recruitment and any environmental
covariate(s) using the prepare_wham_input() function. WHAM
provides 4 options for recruitment (recruit_model):
- random walk,
- random about mean,
- Beverton-Holt, and
- Ricker.
The environmental covariate options are fed to
prepare_wham_input() as a list, ecov:
m=1 # example for first model
ecov <- list(
label = "CPI",
mean = as.matrix(env.dat$CPI),
logsigma = as.matrix(log(env.dat$CPI_sigma)),
year = env.dat$Year,
use_obs = matrix(1, ncol=1, nrow=dim(env.dat)[1]), # use all obs (all = 1)
lag = 1, # CPI in year t affects recruitment in year t+1
process_model = 'rw', # "rw" or "ar1"
where = "none",
how = 0) # 0 = no effect, 1 = controlling, 2 = limitingThere are currently 2 options for the ecov process model
(ecov$process_model): 1) random walk ('rw'),
and 2) autoregressive ('ar1'). We must next specify
where the ecov affects the population;
here it is via recruitment (ecov$where = "recruit") as
opposed to another process like catchability, mortality, maturity, etc.
The options for how the ecov affects
recruitment (ecov$how) follow Iles
and Beverton (1998) and Xu et
al. (2018):
- “controlling” (dens-indep mortality),
- “limiting” (carrying capacity, e.g.
ecovdetermines amount of suitable habitat), - “lethal” (threshold, i.e. R –> 0 at some
ecovvalue),
- “masking” (metabolic/growth,
ecovdecreases dR/dS), and - “directive” (e.g. behavioral).
Finally, we specify the lag at which CPI affects
recruitment (ecov$lag = 1, i.e. CPI in year t
affects recruitment in year t + 1).
You can set ecov = NULL to fit the model without
environmental covariate data, but note that here we fit the
ecov data even for models without an ecov
effect on recruitment (m1 and m3) so that we
can compare them via AIC (need to have the same data in the likelihood).
We accomplish this by setting ecov$how = 0.
Options are described in the prepare_wham_input() help
page. Not all ecov$how options are implemented for every
recruitment model.
?prepare_wham_input3. Run the models
for(m in 1:n.mods){
# set up environmental covariate data and model options
ecov <- list(
label = "CPI",
mean = as.matrix(env.dat$CPI),
logsigma = as.matrix(log(env.dat$CPI_sigma)),
year = env.dat$Year,
use_obs = matrix(1, ncol=1, nrow=dim(env.dat)[1]), # use all obs (=1)
lag = 1, # CPI in year t affects recruitment in year t+1
process_model = df.mods$ecov_process[m], # "rw" or "ar1"
where = c("none","recruit")[as.logical(df.mods$ecov_how[m])+1], # CPI affects recruitment
how = df.mods$ecov_how[m]) # 0 = no effect (but still fit Ecov to compare AIC)
# (not used in this vignette) can set ecov = NULL to fit model without ecov data
if(is.na(df.mods$ecov_process[m])) ecov = NULL
# generate wham input from ASAP3 and ecov data
input <- prepare_wham_input(asap3, recruit_model = df.mods$Recruitment[m],
model_name = "Ex 2: SNEMA Yellowtail Flounder with CPI effects on R",
ecov = ecov,
NAA_re = list(sigma="rec+1", cor="iid"),
age_comp = "logistic-normal-pool0") # logistic normal pool 0 obs
# Selectivity = logistic, not age-specific as in ex1
# 2 pars per block instead of n.ages
# sel pars of indices 4/5 fixed at 1.5, 0.1 (specified via neg phase in ex2_SNEMAYT.dat)
input$par$logit_selpars[1:4,7:8] <- 0 # last 2 rows will not be estimated (mapped to NA)
# Fit model
mod <- fit_wham(input, do.retro=TRUE, do.osa=TRUE)
# Save model
saveRDS(mod, file=paste0(df.mods$Model[m],".rds"))
# Plot output in new subfolder
plot_wham_output(mod=mod, dir.main=file.path(getwd(),df.mods$Model[m]), out.type='html')
}4. Check for convergence
Collect all models into a list.
We need to check that the models converged. The maximum gradient
should be < 1e-6 and SE estimates should be calculable
(invertible Hessian, TMB::sdreport() succeeds). Here,
m6 did not have a positive definite Hessian.
vign2_conv <- lapply(mods, function(x) capture.output(check_convergence(x)))
for(m in 1:n.mods) cat(paste0("Model ",m,":"), vign2_conv[[m]], "", sep='\n')#> Model 1:
#> stats:nlminb thinks the model has converged: mod$opt$convergence == 0
#> Maximum gradient component: 3.41e-10
#> Max gradient parameter: F_devs
#> TMB:sdreport() was performed successfully for this model
#>
#> Model 2:
#> stats:nlminb thinks the model has converged: mod$opt$convergence == 0
#> Maximum gradient component: 5.95e-11
#> Max gradient parameter: logit_selpars
#> TMB:sdreport() was performed successfully for this model
#>
#> Model 3:
#> stats:nlminb thinks the model has converged: mod$opt$convergence == 0
#> Maximum gradient component: 6.15e-11
#> Max gradient parameter: log_F1
#> TMB:sdreport() was performed successfully for this model
#>
#> Model 4:
#> stats:nlminb thinks the model has converged: mod$opt$convergence == 0
#> Maximum gradient component: 6.00e-10
#> Max gradient parameter: logit_selpars
#> TMB:sdreport() was performed successfully for this model
#>
#> Model 5:
#> stats:nlminb thinks the model has converged: mod$opt$convergence == 0
#> Maximum gradient component: 3.10e-10
#> Max gradient parameter: logit_selpars
#> TMB:sdreport() was performed successfully for this model
#>
#> Model 6:
#> stats:nlminb thinks the model has converged: mod$opt$convergence == 0
#> Maximum gradient component: 1.41e-09
#> Max gradient parameter: logit_selpars
#> TMB:sdreport() was performed successfully for this model
#>
#> Model 7:
#> stats:nlminb thinks the model has converged: mod$opt$convergence == 0
#> Maximum gradient component: 8.49e-11
#> Max gradient parameter: logit_selpars
#> TMB:sdreport() was performed successfully for this model5. Compare models
Let’s first make the results table prettier.
df.mods$Recruitment <- dplyr::recode(df.mods$Recruitment, `2`='Random', `3`='Bev-Holt', `4`='Ricker')
df.mods$ecov_how <- dplyr::recode(df.mods$ecov_how, `0`='---',`1`='Controlling', `2`='Limiting', `4`='Masking')Now get the convergence information.
opt_conv = 1-sapply(mods, function(x) x$opt$convergence)
ok_sdrep = sapply(mods, function(x) if(x$na_sdrep==FALSE & !is.na(x$na_sdrep)) 1 else 0)
df.mods$conv <- as.logical(opt_conv)
df.mods$pdHess <- as.logical(ok_sdrep)
df.mods$NLL <- sapply(mods, function(x) round(x$opt$objective,3))Only calculate AIC and Mohn’s rho for converged models.
not_conv <- !df.mods$conv | !df.mods$pdHess
mods2 <- mods
mods2[not_conv] <- NULL
df.aic.tmp <- as.data.frame(compare_wham_models(mods2, table.opts=list(sort=FALSE, calc.rho=T))$tab)
df.aic <- df.aic.tmp[FALSE,]
ct = 1
for(i in 1:n.mods){
if(not_conv[i]){
df.aic[i,] <- rep(NA,5)
} else {
df.aic[i,] <- df.aic.tmp[ct,]
ct <- ct + 1
}
}
df.mods <- cbind(df.mods, df.aic)
df.mods <- df.mods[order(df.mods$dAIC, na.last=TRUE),]
df.mods[is.na(df.mods$AIC), c('dAIC','AIC','rho_R','rho_SSB','rho_Fbar')] <- "---"
rownames(df.mods) <- NULLPrint and save the results table. m5 has the lowest AIC
(Bev-Holt recruitment, CPI modeled as AR1, limiting effect of CPI on
recruitment).
save("df.mods", file="vign2_res.RData")
df.mods#> Model Recruitment Ecov_process Ecov_how conv pdHess NLL dAIC AIC
#> 1 m6 Bev-Holt ar1 Controlling TRUE TRUE -829.795 0.0 -1519.6
#> 2 m5 Bev-Holt ar1 Limiting TRUE TRUE -829.414 0.8 -1518.8
#> 3 m7 Ricker ar1 Controlling TRUE TRUE -829.124 1.4 -1518.2
#> 4 m4 Bev-Holt rw Limiting TRUE TRUE -818.699 20.2 -1499.4
#> 5 m2 Random rw Controlling TRUE TRUE -815.937 23.7 -1495.9
#> 6 m3 Bev-Holt rw --- TRUE TRUE -813.130 29.3 -1490.3
#> 7 m1 Random rw --- TRUE TRUE -808.943 35.7 -1483.9
#> rho_R rho_SSB rho_Fbar
#> 1 0.1897 0.0653 -0.1016
#> 2 0.1963 0.0690 -0.1052
#> 3 0.1885 0.0622 -0.0987
#> 4 0.1820 0.0670 -0.1042
#> 5 0.1974 0.0718 -0.1102
#> 6 0.2121 0.0679 -0.1033
#> 7 0.2308 0.0714 -0.11066. Results
There are 3 options for plotting WHAM output. The default
(out.type='html') creates and opens an HTML file with plots
organized into tabs (code modified from r4ss::SS_html()):
# save output plots in subfolder for each model
for(m in 1:n.mods) plot_wham_output(mod=mods[[m]], dir.main=file.path(getwd(), df.mods$Model[m]), out.type='html')Cold Pool Index (CPI)
Models that included an effect of the Cold Pool Index on recruitment
were strongly supported by AIC over models without CPI effects
(m2 and m4-7 lower AIC than m1
and m3). Note that we can compare models with and without a
CPI effect on recruitment using AIC because we also fit the CPI data in
the models without the effect (m1 and m3).
Comparing m4 and m5 demonstrates that the
CPI was best modeled as an AR1 process (m5) instead of a
random walk (m4), since this was the only difference
between the two models and m5 had lower AIC. In addition,
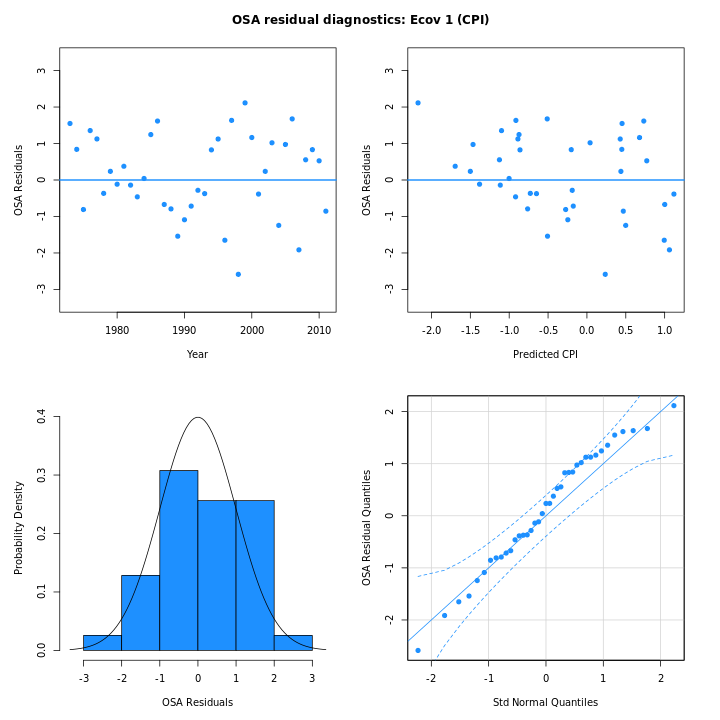
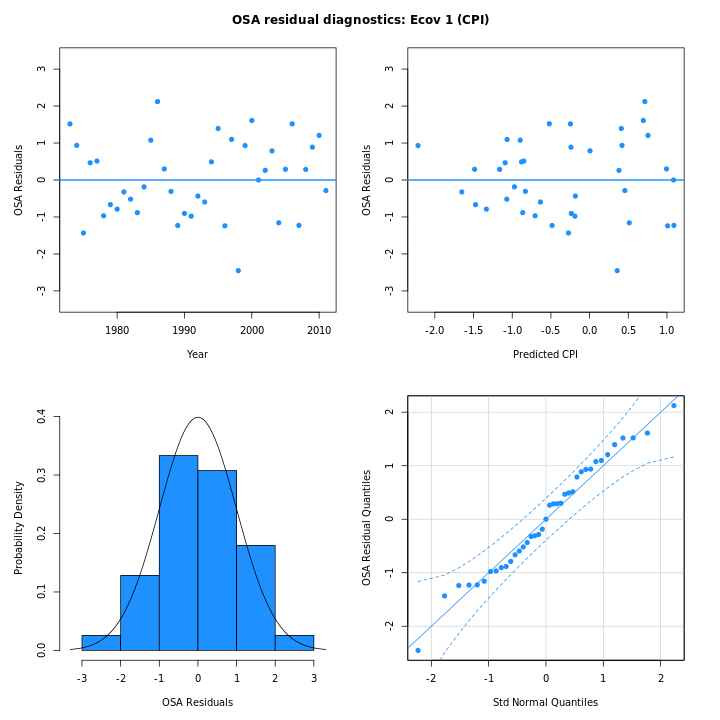
the one-step-ahead residuals for the CPI from m5 (right)
are smaller and conform more closely to a normal distribution than in
m4 (left):
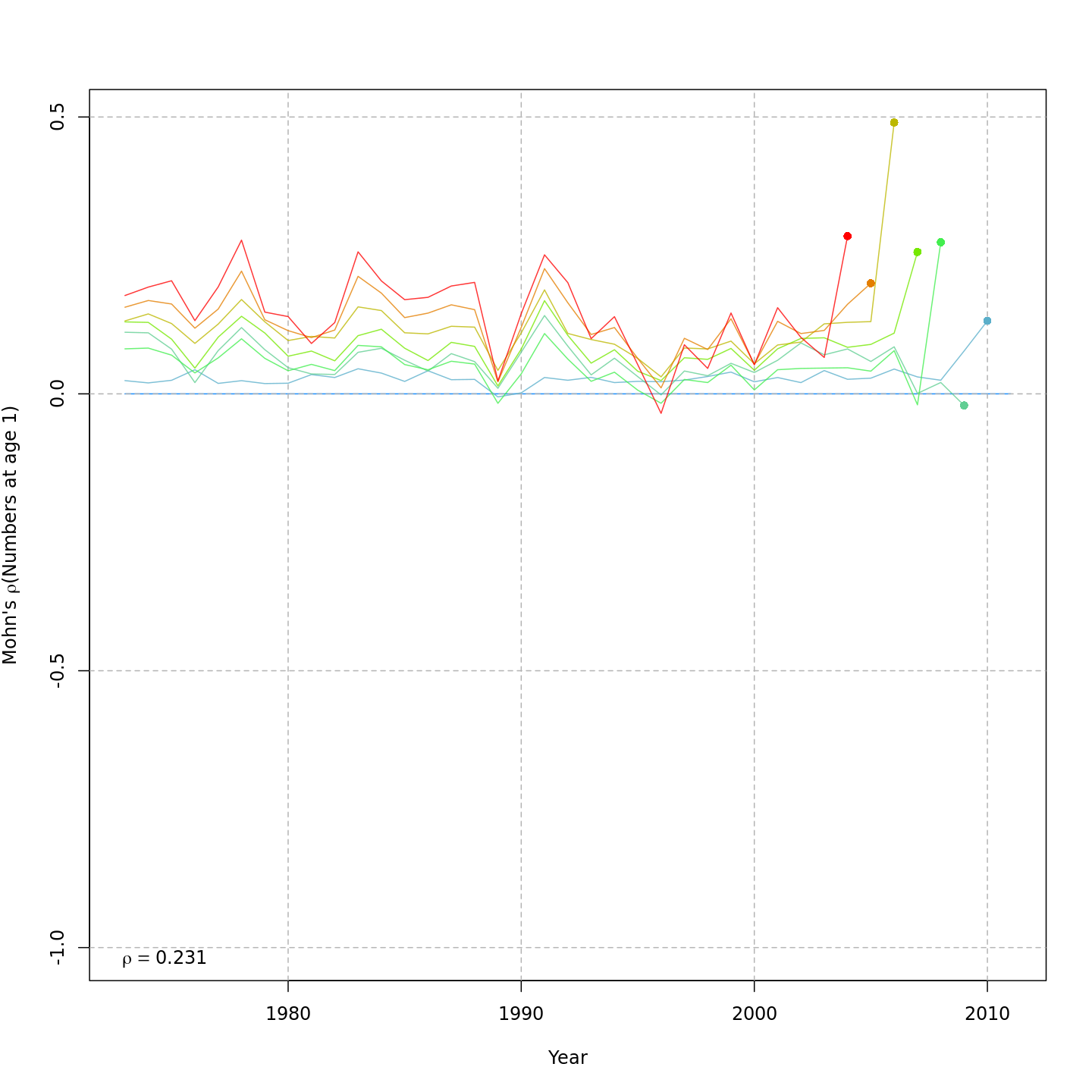
Compared to the base model (m1, left), the best model
that included CPI and SSB effects on recruitment, (m5,
right) reduced the retrospective pattern in recruitment, \(\rho_R\), very slightly from 0.231 to
0.196.
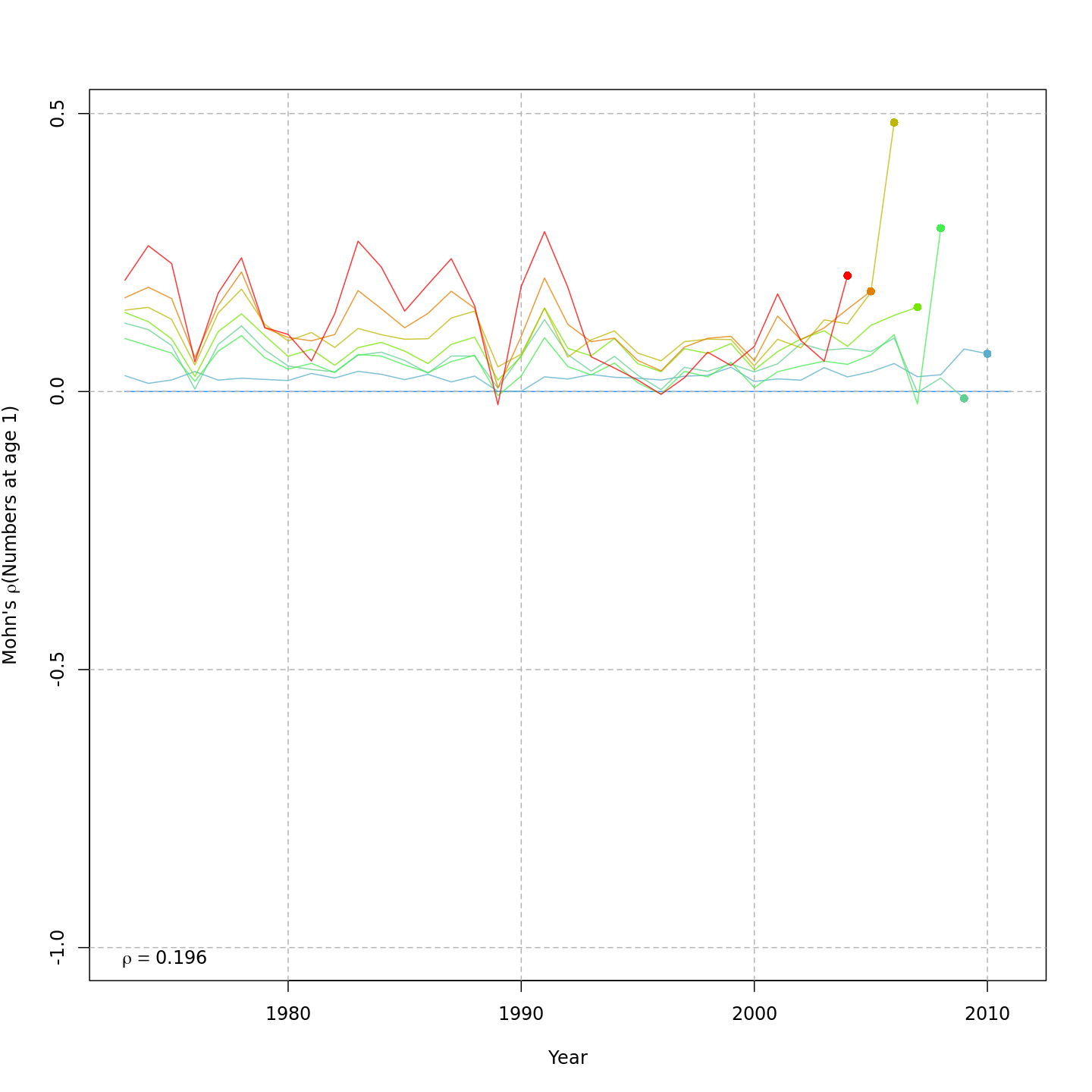
Recruitment
Beverton-Holt recruitment was preferred over random (m4
lower AIC than m2) and weakly preferred over Ricker
(m5 lower AIC than m7). Models that included
both Bev-Holt and CPI effects on recruitment had lower AIC than the
model with Bev-Holt but without the CPI (m4
vs. m3). Adding the CPI effect to the Bev-Holt explains
some of the variability around the stock-recruit curve, which resulted
in m4 (right) estimating lower \(\sigma^2_R\) than m3
(left).
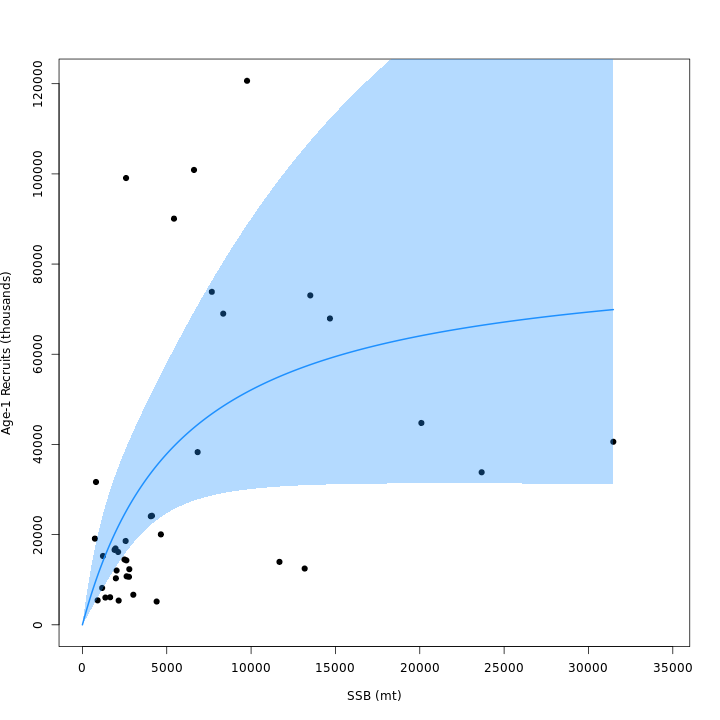
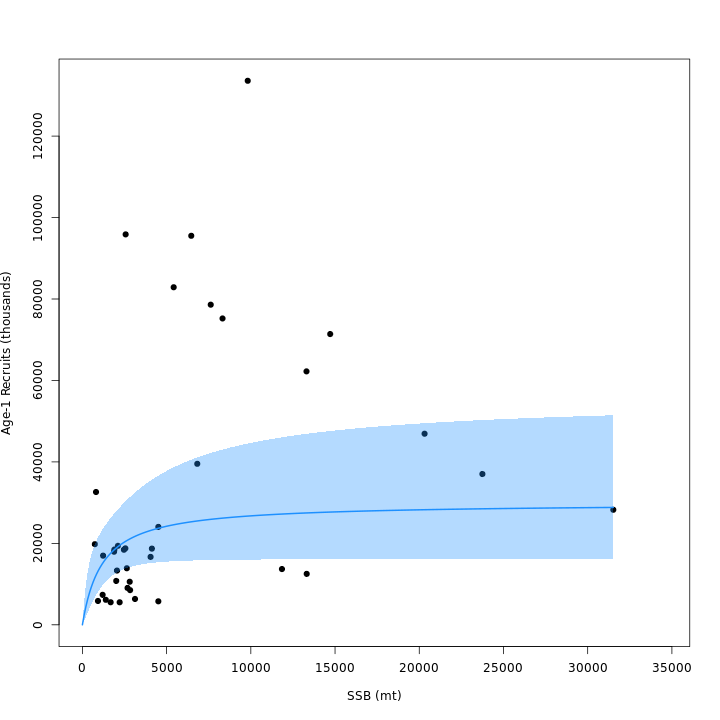
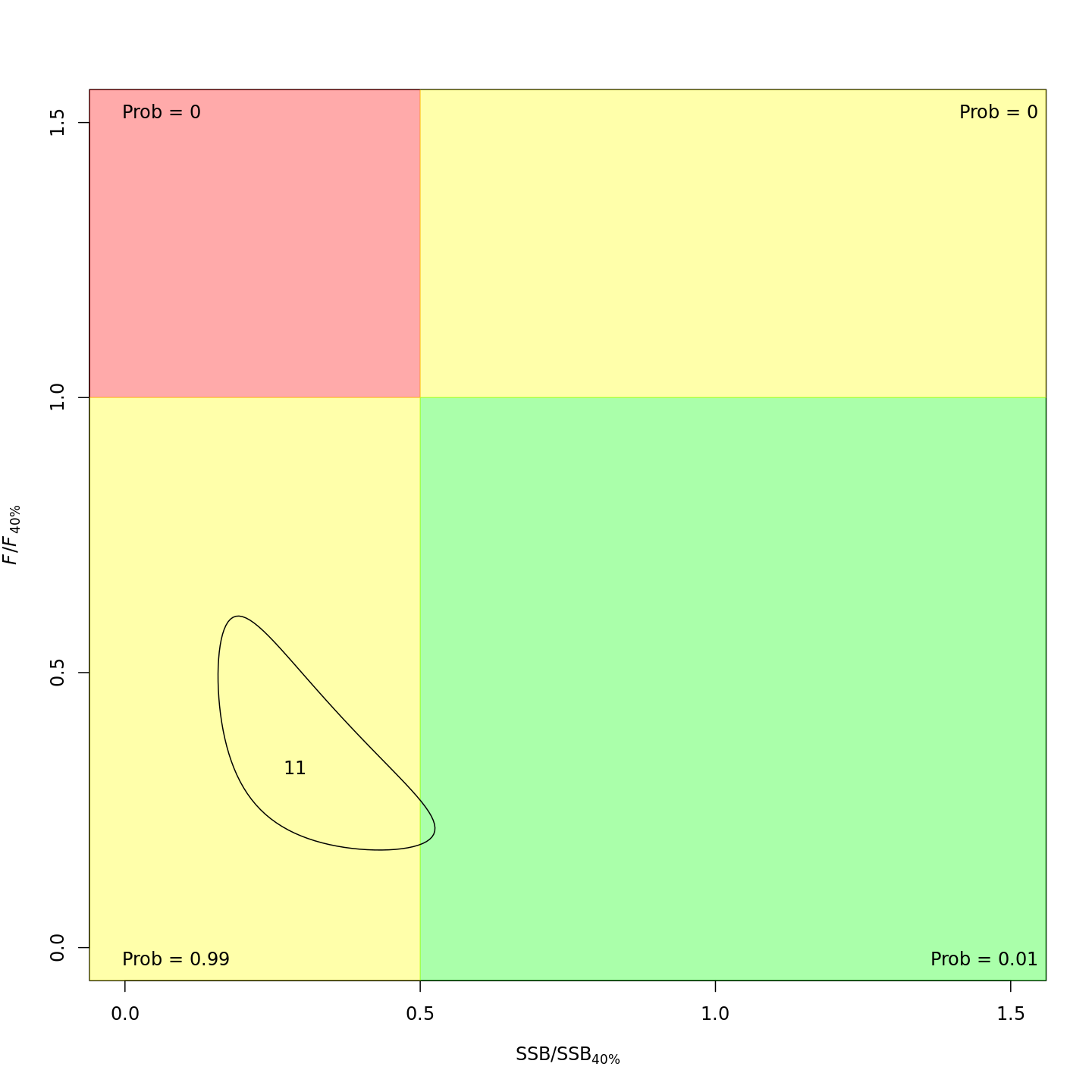
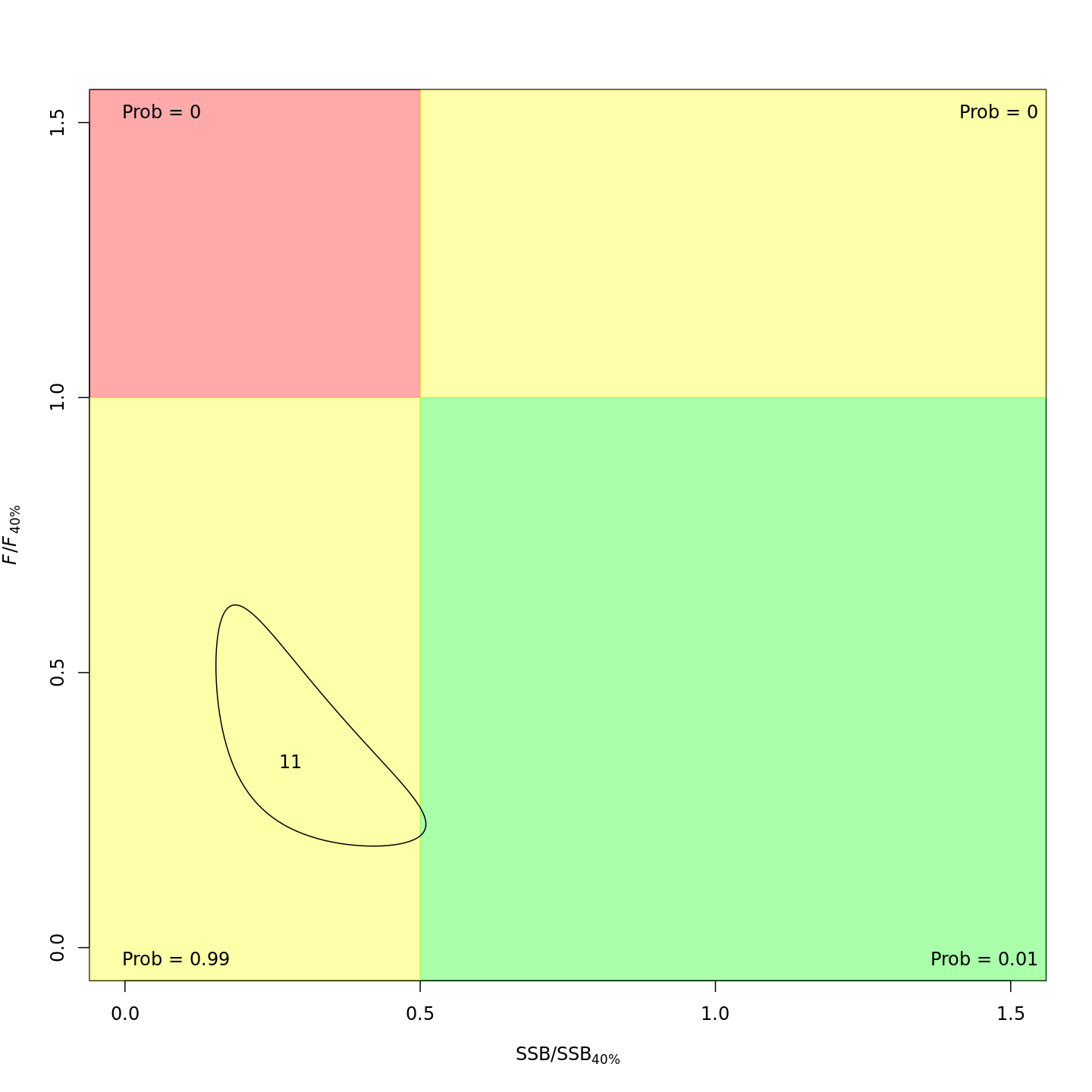
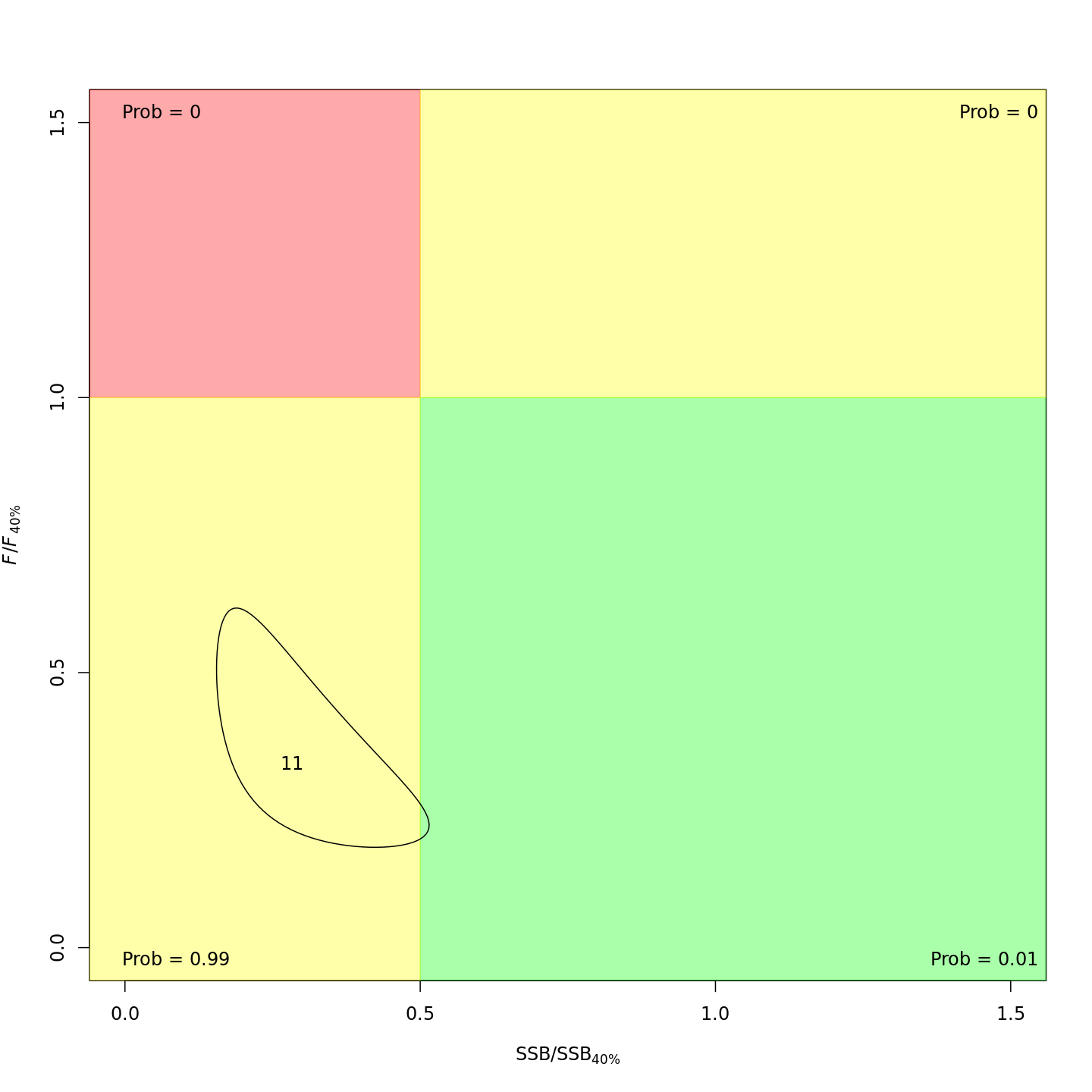
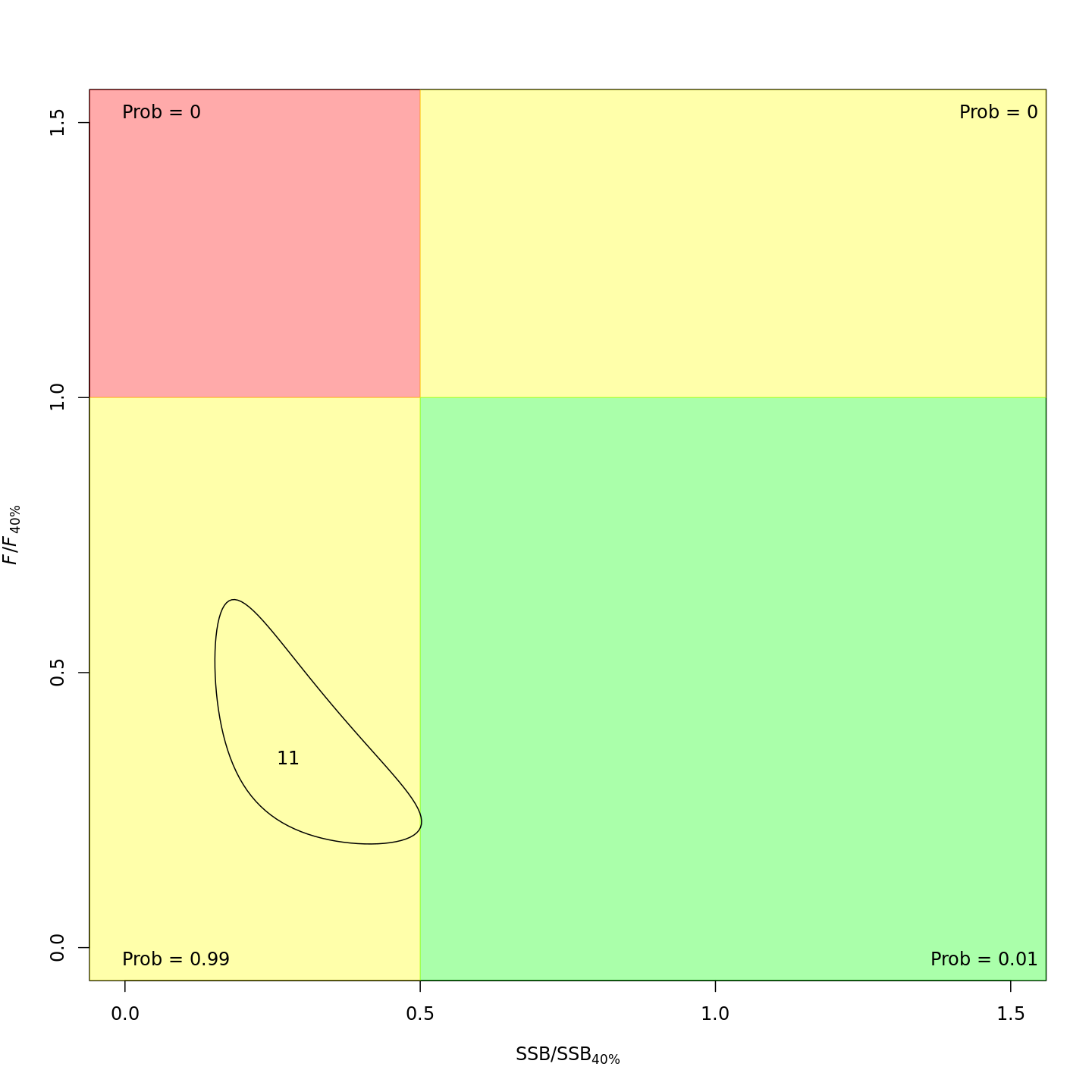
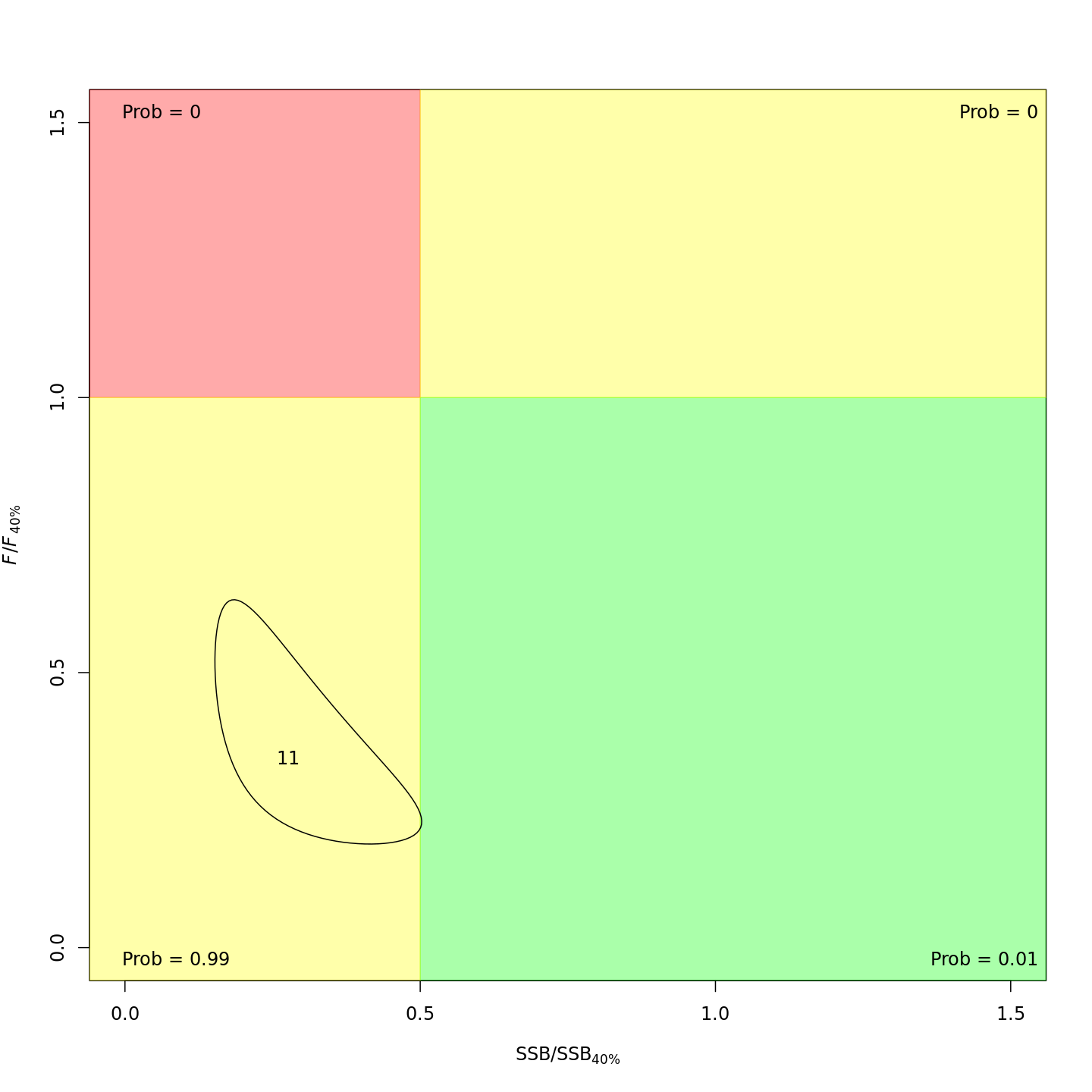
Stock status
Whether or not to include a stock-recruit function and/or the CPI did not have a great influence on estimated stock status. Specifically, the models hardly differed in their estimation of the probability that the stock was overfished, \(Pr[SSB < 0.5 \: SSB_{40\%}]\). All models estimated with 100% probability that the stock was not experiencing overfishing in 2011, \(F < F_{40\%}\).