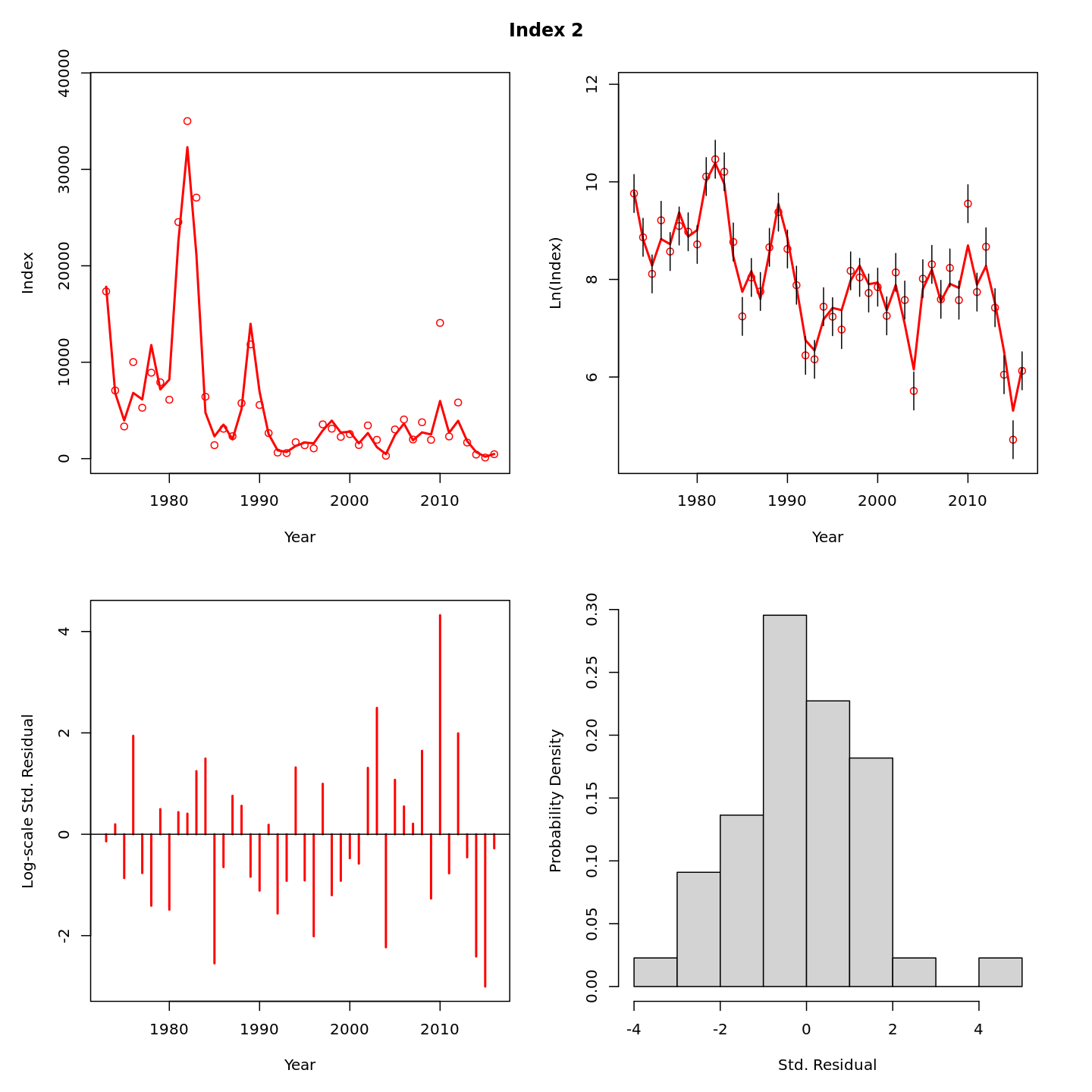
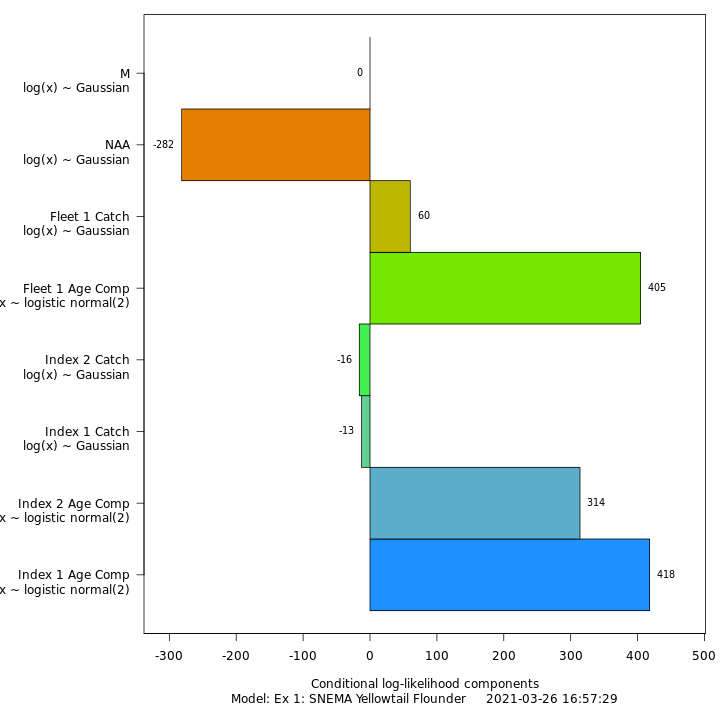
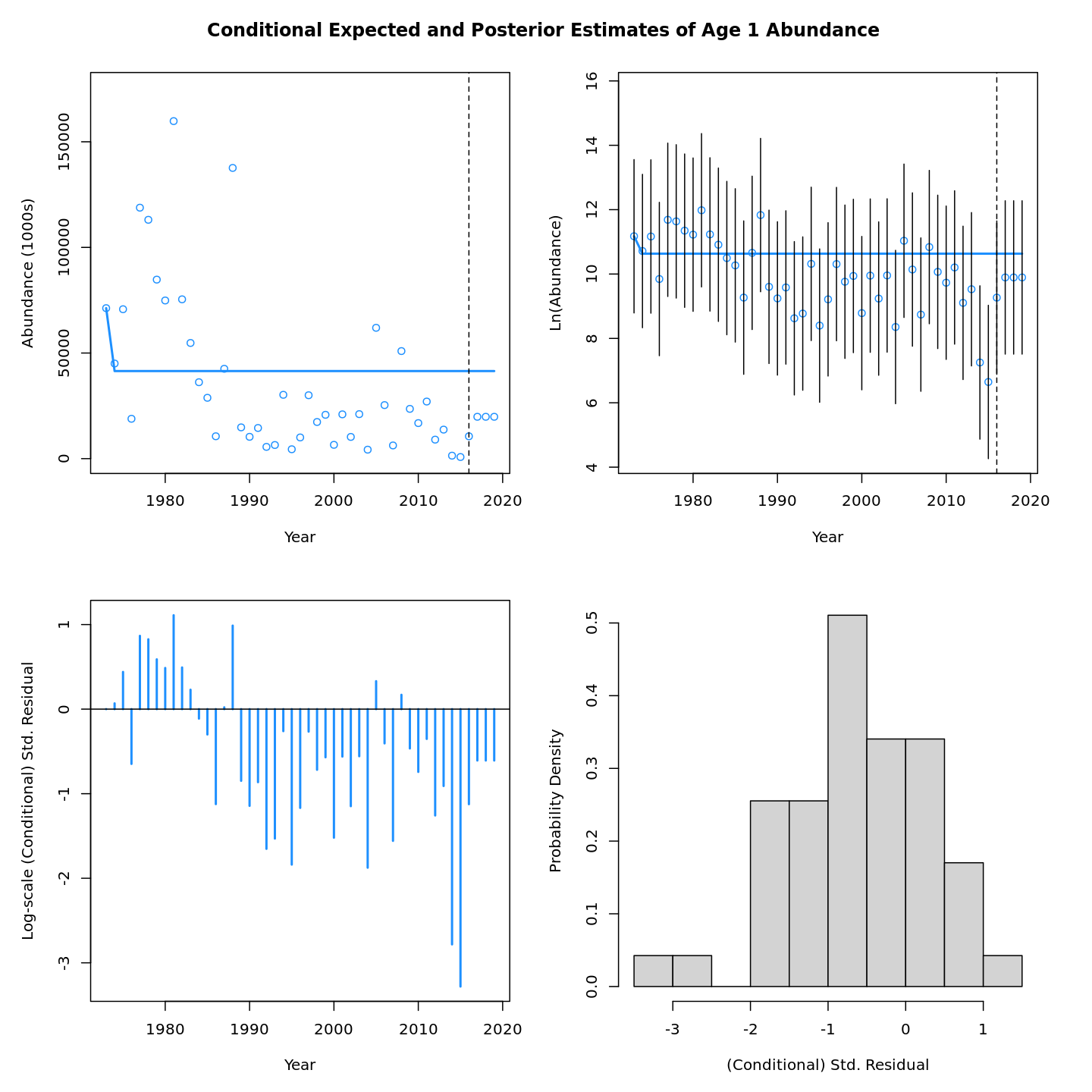
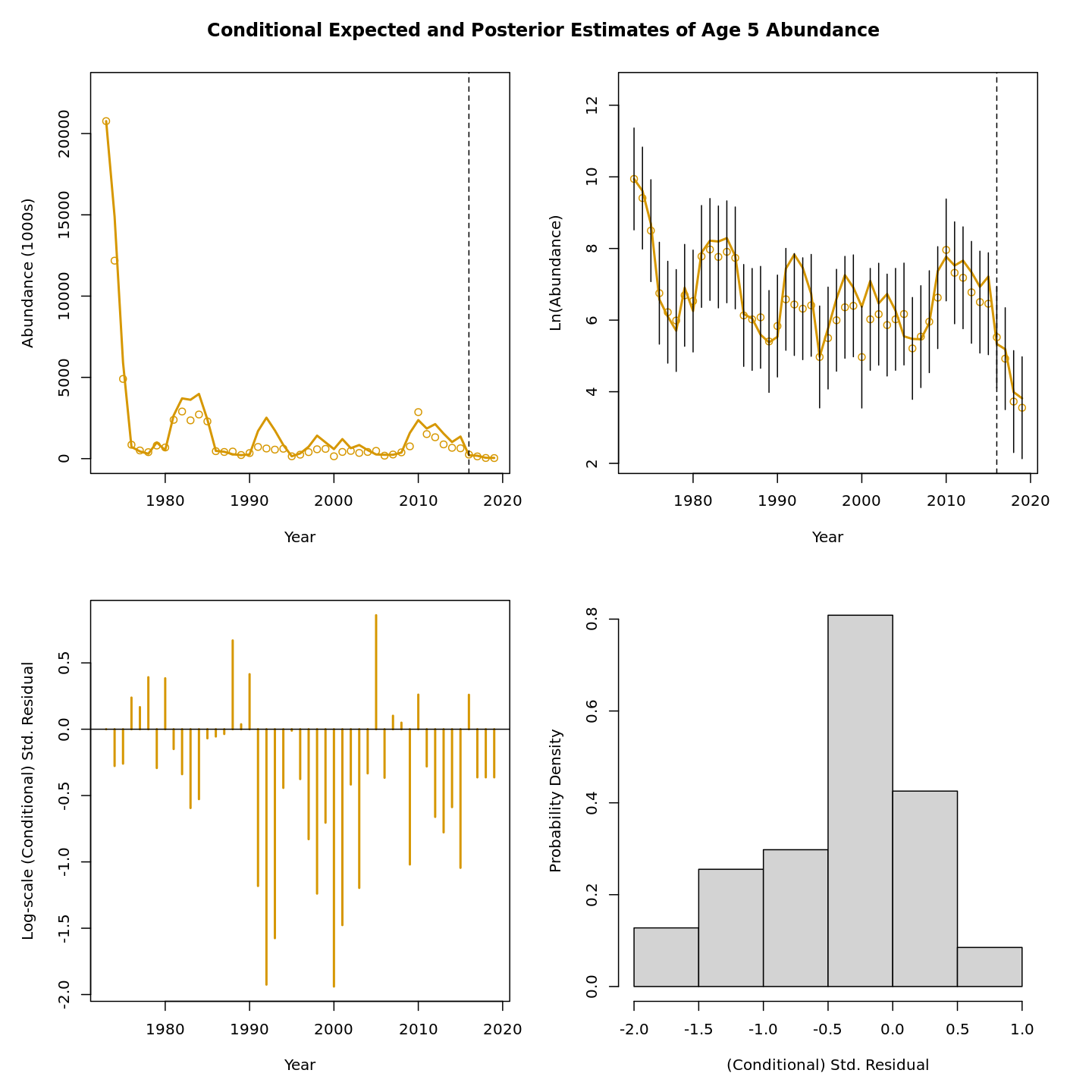
In this vignette we walk through an example using the
wham (WHAM = the Woods Hole Assessment Model) package to
run a state-space age-structured stock assessment model. WHAM is a
generalization of code written for Miller et al. (2016),
and in this example we apply WHAM to the same stock as in Miller et
al. (2016), Southern New England / Mid-Atlantic Yellowtail Flounder.
Here, we demonstrate the basic wham workflow:
Load
whamand data-
Specify several (slightly different) models:
m1: statistical catch-at-age (SCAA) model, but with recruitment estimated as random effects; multinomial age-compositions
m2: as m1, but with logistic normal age-compositions
m3: full state-space model (numbers at all ages are random effects), multinomial age-compositions
m4: full state-space model, logistic normal age-compositions
Fit models and check for convergence
Compare models by AIC and Mohn’s rho (retrospective analysis)
Review plots of input data, diagnostics, and results.
1. Load data
We assume you have already read the Introduction and installed
wham and its dependencies. If not, you should be able to
just run
devtools::install_github("timjmiller/wham", dependencies=TRUE).
Open R and load the wham package:
For a clean, runnable .R script, look at
ex1_basics.R in the example_scripts folder of
the wham package install:
wham.dir <- find.package("wham")
file.path(wham.dir, "example_scripts")
#> [1] "C:/Users/timothy.j.miller/AppData/Local/R/win-library/4.2/wham/example_scripts"You can run this entire example script with (2.0 min runtime):
write.dir <- "choose/where/to/save/output" # otherwise will be saved in working directory
source(file.path(wham.dir, "example_scripts", "ex1_basics.R"))Let’s create a directory for this analysis:
# choose a location to save output
write.dir <- "/path/to/save/output" # modify
dir.create(write.dir)
setwd(write.dir)WHAM was built by modifying the ADMB-based ASAP model code (Legault and
Restrepo 1999), and is designed to take an ASAP3 .dat file as input.
We generally assume in wham that you have an existing ASAP3
.dat file. If you are not familiar with ASAP3 input files, see the ASAP
documentation
and code. For this
vignette, an example ASAP3 input file is provided.
Copy ex1_SNEMAYT.dat to our analysis directory:
wham.dir <- find.package("wham")
file.copy(from=file.path(wham.dir,"extdata","ex1_SNEMAYT.dat"), to=write.dir, overwrite=FALSE)Confirm you are in the working directory and it has the
ex1_SNEMAYT.dat file:
list.files()
#> [1] "age_comp_likelihood_test_values.rds" "BASE_3"
#> [3] "CPI.csv" "ex1_SNEMAYT.dat"
#> [5] "ex1_test_results.rds" "ex11_tests.rds"
#> [7] "ex2_SNEMAYT.dat" "ex2_test_results.rds"
#> [9] "ex4_test_results.rds" "ex5_summerflounder.dat"
#> [11] "ex5_test_results.rds" "ex6_test_results.rds"
#> [13] "ex7_SNEMAYT.dat" "GSI.csv"Read the ASAP3 .dat file into R:
asap3 <- read_asap3_dat("ex1_SNEMAYT.dat")2. Specify model
We use the prepare_wham_input() function to specify the
model name and any settings that differ from the ASAP3 file. Our first
model will use:
- recruitment model: random about mean, no S-R function
(
recruit_model = 2) - recruitment deviations: independent random effects
(
NAA_re = list(sigma="rec", cor="iid")) - selectivity: age-specific (fix sel=1 for ages 4-5 in fishery, age 4 in index1, and ages 2-4 in index2)
input1 <- prepare_wham_input(asap3, recruit_model=2, model_name="Ex 1: SNEMA Yellowtail Flounder",
selectivity=list(model=rep("age-specific",3),
re=rep("none",3),
initial_pars=list(c(0.5,0.5,0.5,1,1,0.5),c(0.5,0.5,0.5,1,0.5,0.5),c(0.5,1,1,1,0.5,0.5)),
fix_pars=list(4:5,4,2:4)),
NAA_re = list(sigma="rec", cor="iid"))The stock-recruit model options in WHAM are:
- 1 = random walk,
- 2 = random about mean (default),
- 3 = Beverton-Holt, and
- 4 = Ricker
Note that the parameterization of age-specific selectivity is
specific to SNEMA yellowtail flounder. We will use age-specific
selectivity parameters for the first three selectivity blocks.
Selectivity blocks define a selectivity configuration that can be used
in one or more fleets or surveys over any set of years in the model.
prepare_wham_input() fixes age-specific parameters at zero
if there are any age classes without any observations where the
selectivity block is applied. The function configures all other
age-specific parameters to be estimated. Generally there may be
confounding of the selectivity parameters with either fully-selected
fishing mortality (for fleets) or catchability (for surveys), so
selectivity parameters for one or more ages would need to be fixed to
allow convergence. An initial fit with all selectivity parameters freely
estimated can be useful in determining which age(s) to fix selectivity
at 1. In the code above, we have already determined the ages to fix
selectivity at 1 (ages 4-5 in fishery, age 4 in index 1, and ages 2-4 in
index 2). If you are interested in more details and options for
selectivity in WHAM, see Example
4 and prepare_wham_input().
3. Fit model and check for convergence
m1 <- fit_wham(input1, do.osa = F) # turn off OSA residuals to save time in exBy default, fit_wham() uses 3 extra Newton steps to
reduce the absolute value of the gradient (n.newton = 3)
and estimates standard errors for derived parameters
(do.sdrep = TRUE). fit_wham() also does a
retrospective analysis with 7 peels by default
(do.retro = TRUE, n.peels = 7). For more
details, see fit_wham().
We need to check that m1 converged
(m1$opt$convergence should be 0, and the maximum absolute
value of the gradient vector should be < 1e-06). Convergence issues
may indicate that a model is misspecified or overparameterized. To help
diagnose these problems, fit_wham() includes a
do.check option to run an internal
check_estimability function originally
written by Jim Thorson. do.check = FALSE by default. To
turn on, set do.check = TRUE. See
fit_wham().
#> stats:nlminb thinks the model has converged: mod$opt$convergence == 0
#> Maximum gradient component: 1.34e-12
#> Max gradient parameter: logit_q
#> TMB:sdreport() was performed successfully for this modelFit models m2-m4
The second model, m2, is like the first, but changes all
the age composition likelihoods from multinomial to logistic normal
(treating 0 observations as missing):
input2 <- prepare_wham_input(asap3, recruit_model=2, model_name="Ex 1: SNEMA Yellowtail Flounder",
selectivity=list(model=rep("age-specific",3),
re=rep("none",3),
initial_pars=list(c(0.5,0.5,0.5,1,1,0.5),c(0.5,0.5,0.5,1,0.5,0.5),c(0.5,1,1,1,0.5,0.5)),
fix_pars=list(4:5,4,2:4)),
NAA_re = list(sigma="rec", cor="iid"),
age_comp = "logistic-normal-miss0")
m2 <- fit_wham(input2, do.osa = F) # turn off OSA residuals to save time in exCheck that m2 converged:
#> stats:nlminb thinks the model has converged: mod$opt$convergence == 0
#> Maximum gradient component: 2.42e-12
#> Max gradient parameter: index_paa_pars
#> TMB:sdreport() was performed successfully for this modelThe third, m3, is a full state-space model where numbers
at all ages are random effects
(NAA_re$sigma = "rec+1"):
input3 <- prepare_wham_input(asap3, recruit_model=2, model_name="Ex 1: SNEMA Yellowtail Flounder",
selectivity=list(model=rep("age-specific",3),
re=rep("none",3),
initial_pars=list(c(0.5,0.5,0.5,1,1,0.5),c(0.5,0.5,0.5,1,0.5,0.5),c(0.5,1,1,1,0.5,0.5)),
fix_pars=list(4:5,4,2:4)),
NAA_re = list(sigma="rec+1", cor="iid"))
m3 <- fit_wham(input3, do.osa = F) # turn off OSA residuals to save time in exCheck that m3 converged:
#> stats:nlminb thinks the model has converged: mod$opt$convergence == 0
#> Maximum gradient component: 1.89e-05
#> Max gradient parameter: log_F1
#> TMB:sdreport() was performed successfully for this modelThe last, m4, is like m3, but again changes
all the age composition likelihoods to logistic normal:
input4 <- prepare_wham_input(asap3, recruit_model=2, model_name="Ex 1: SNEMA Yellowtail Flounder",
selectivity=list(model=rep("age-specific",3),
re=rep("none",3),
initial_pars=list(c(0.5,0.5,0.5,1,1,0.5),c(0.5,0.5,0.5,1,0.5,0.5),c(0.5,1,1,1,0.5,0.5)),
fix_pars=list(4:5,4,2:4)),
NAA_re = list(sigma="rec+1", cor="iid"),
age_comp = "logistic-normal-miss0")
m4 <- fit_wham(input4, do.osa = F) # turn off OSA residuals to save time in exCheck that m4 converged:
#> stats:nlminb thinks the model has converged: mod$opt$convergence == 0
#> Maximum gradient component: 7.24e-11
#> Max gradient parameter: F_devs
#> TMB:sdreport() was performed successfully for this modelStore all models together in one (named) list:
mods <- list(m1=m1, m2=m2, m3=m3, m4=m4)Since the retrospective analyses take a few minutes to run, you may want to save the output for later use:
save("mods", file="ex1_models.RData")4. Compare models
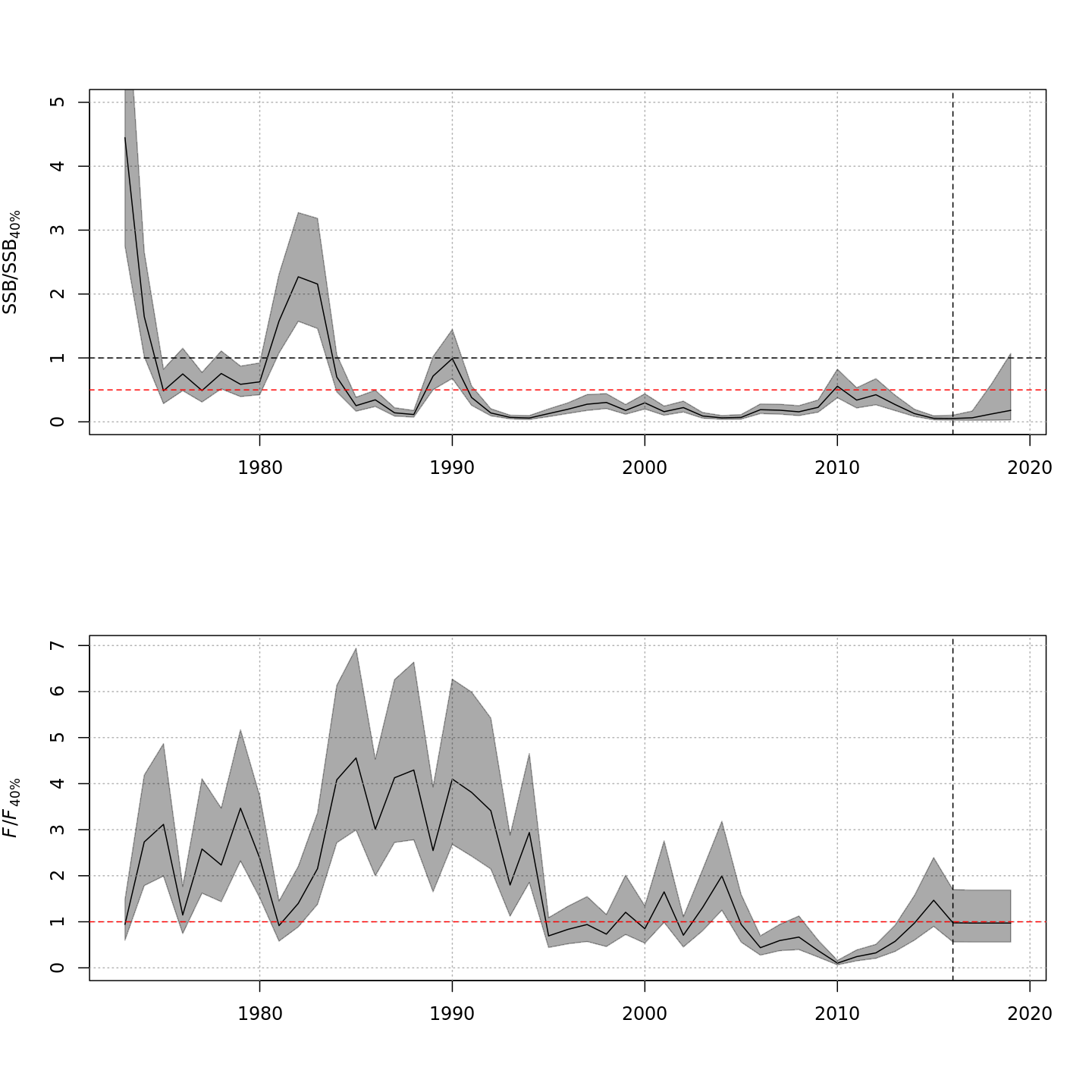
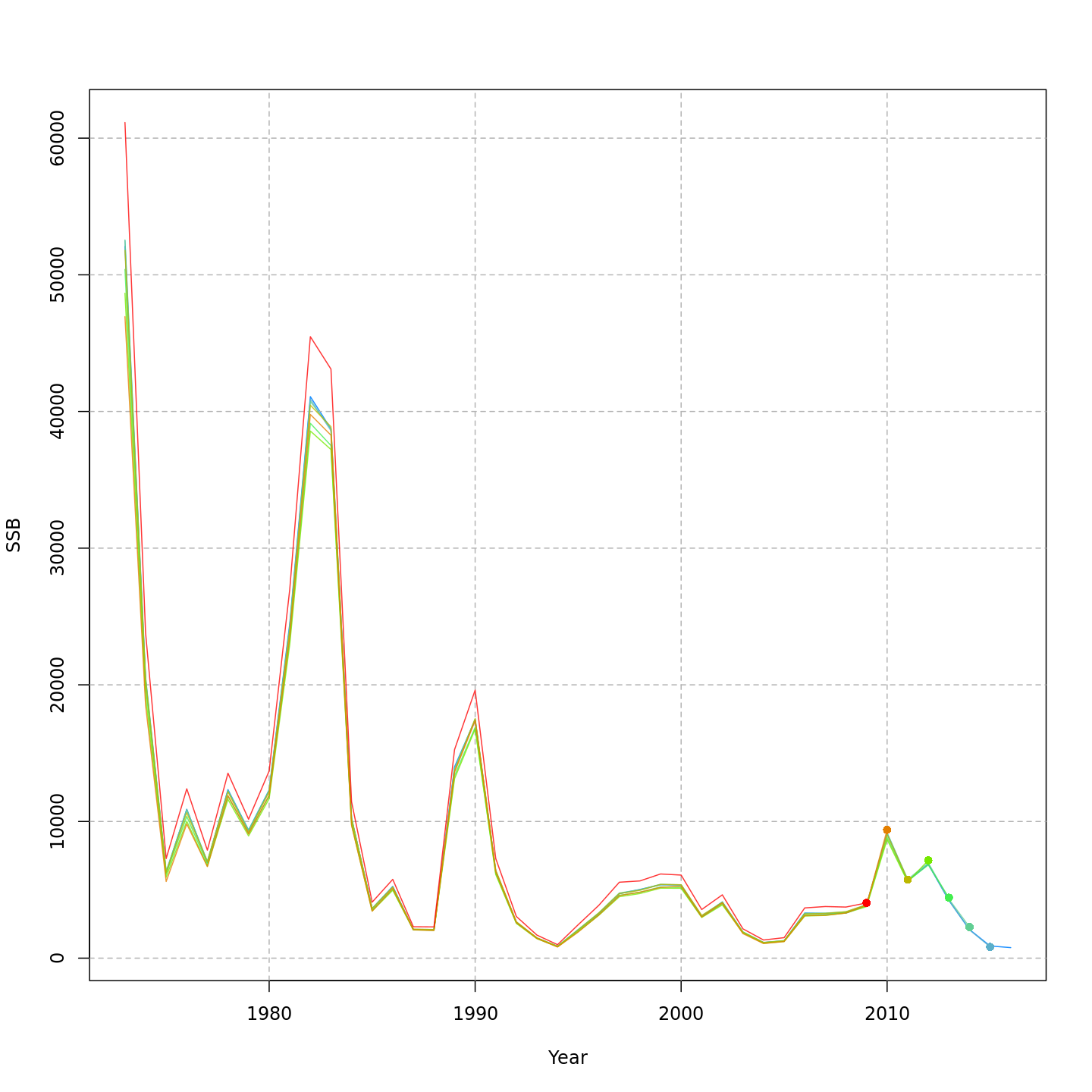
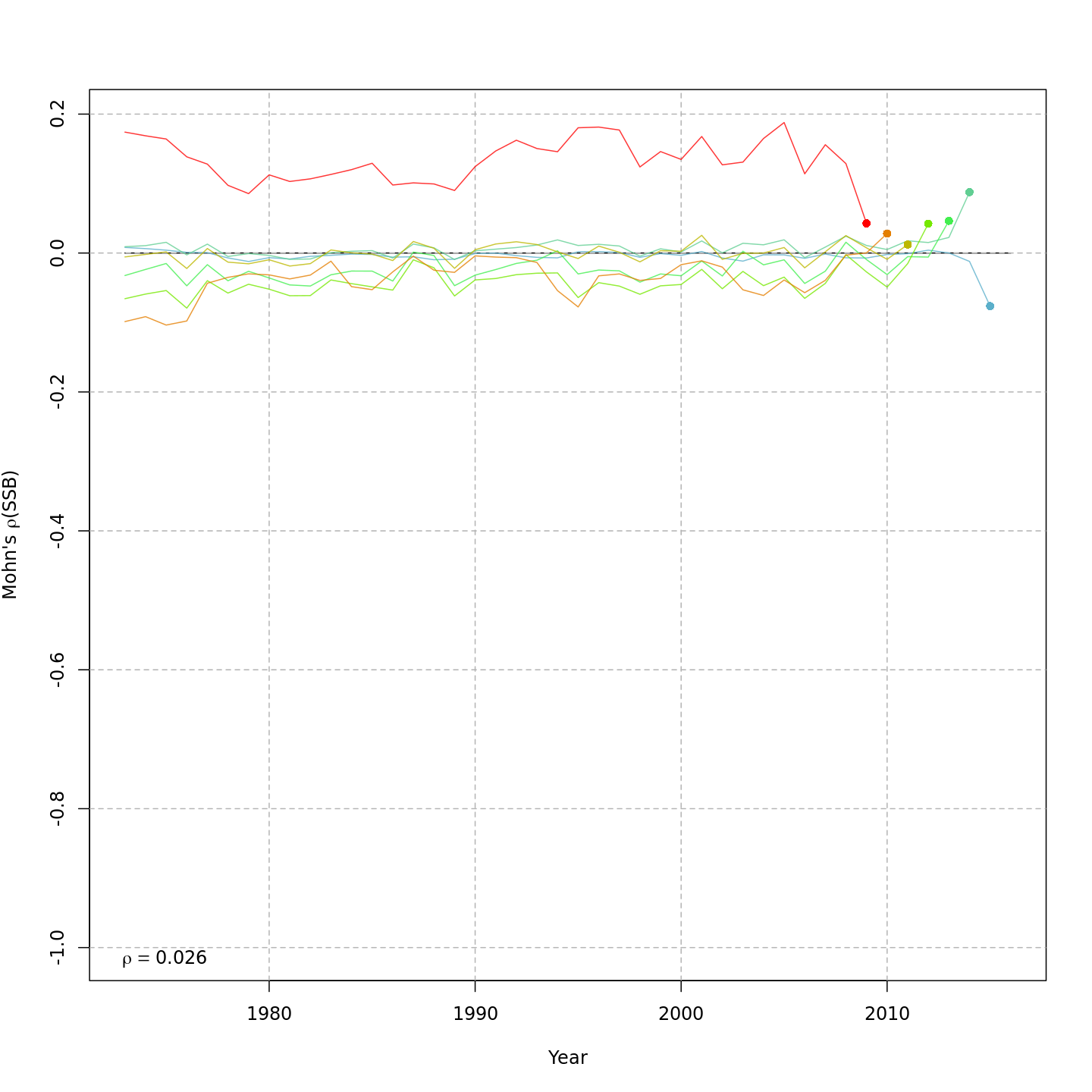
compare_wham_models() will make 1) a table comparing
multiple WHAM model fits using AIC and Mohn’s rho, and 2) plots of key
output (e.g. SSB, F, recruitment, reference points).
res <- compare_wham_models(mods, table.opts=list(fname="ex1_table", sort=TRUE))#> dAIC AIC rho_R rho_SSB rho_Fbar
#> m4 0.0 -1481.5 0.2890 0.0261 -0.0286
#> m2 315.1 -1166.4 5.1190 -0.0205 0.0174
#> m3 5575.9 4094.4 0.1124 0.0053 -0.0056
#> m1 6322.0 4840.5 0.8207 0.1840 -0.1748
res$best#> [1] "m4"By default, compare_wham_models() sorts the model
comparison table with lowest (best) AIC at the top, and saves it as
model_comparison.csv. However, in this example the models
with alternative likelihoods for the age composition observations are
not comparable due to differences in how the observations are defined.
Still, the models that treat the age composition observations in the
same way can be compared to evaluate whether stochasticity in abundances
at age provides better performance, i.e. m1
vs. m3 (both multinomial) and m2
vs. m4 (both logistic normal).
The comparison plots are stored by default in the
compare_png folder.
Project the best model
Let’s do projections for the best model, m4, using the
default settings (see project_wham()):
m4_proj <- project_wham(model=m4)5. Plot input data, diagnostics, and results
There are 3 options for plotting WHAM output. The default
(out.type='png') creates a plots_png directory
with plots organized into subdirectories.
plot_wham_output(mod=m4_proj) # default is pngAnother option is out.type='html', which creates the
same .png files and then opens an HTML file with plots
organized into tabs (code modified from r4ss::SS_html()).
This option will also generate and open html tables of estimates for
fundamental parameters and numbers and fishing mortality at age. On
Windows you may need to use Chrome or Internet Explorer to view the
.html (there have been issues using Firefox on Windows but
not Linux).
plot_wham_output(mod=m4_proj, out.type='html')
Example HTML file created by
plot_wham_output(mod=m4, out.type='html').
Setting out.type='pdf' saves the plots organized into 6
.pdf files corresponding to the tabs in the
.html file (Diagnostics, Input Data, Results, Reference
Points, Retrospective, and Misc). This option will also generate a pdf
of the same tables as those under the html option.
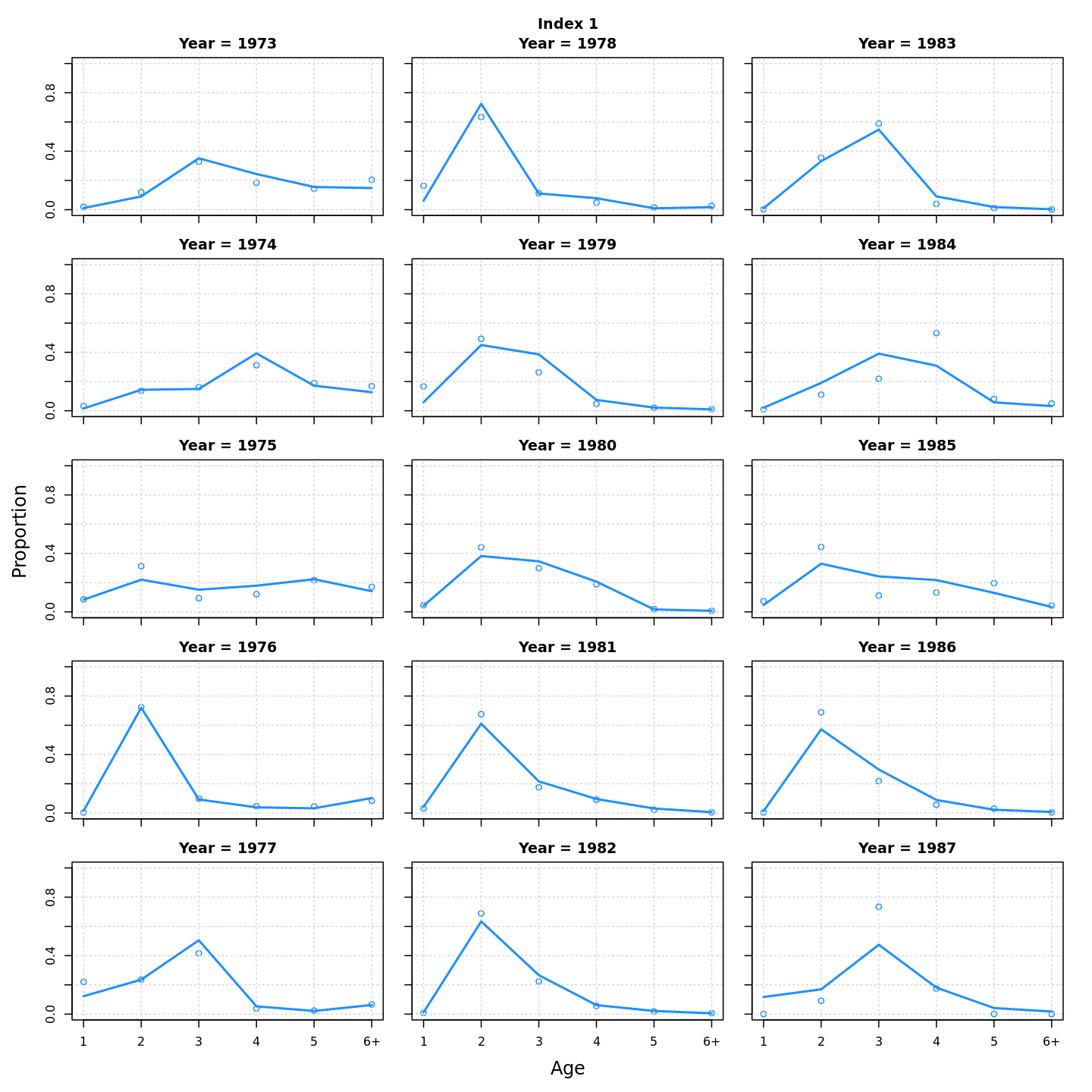
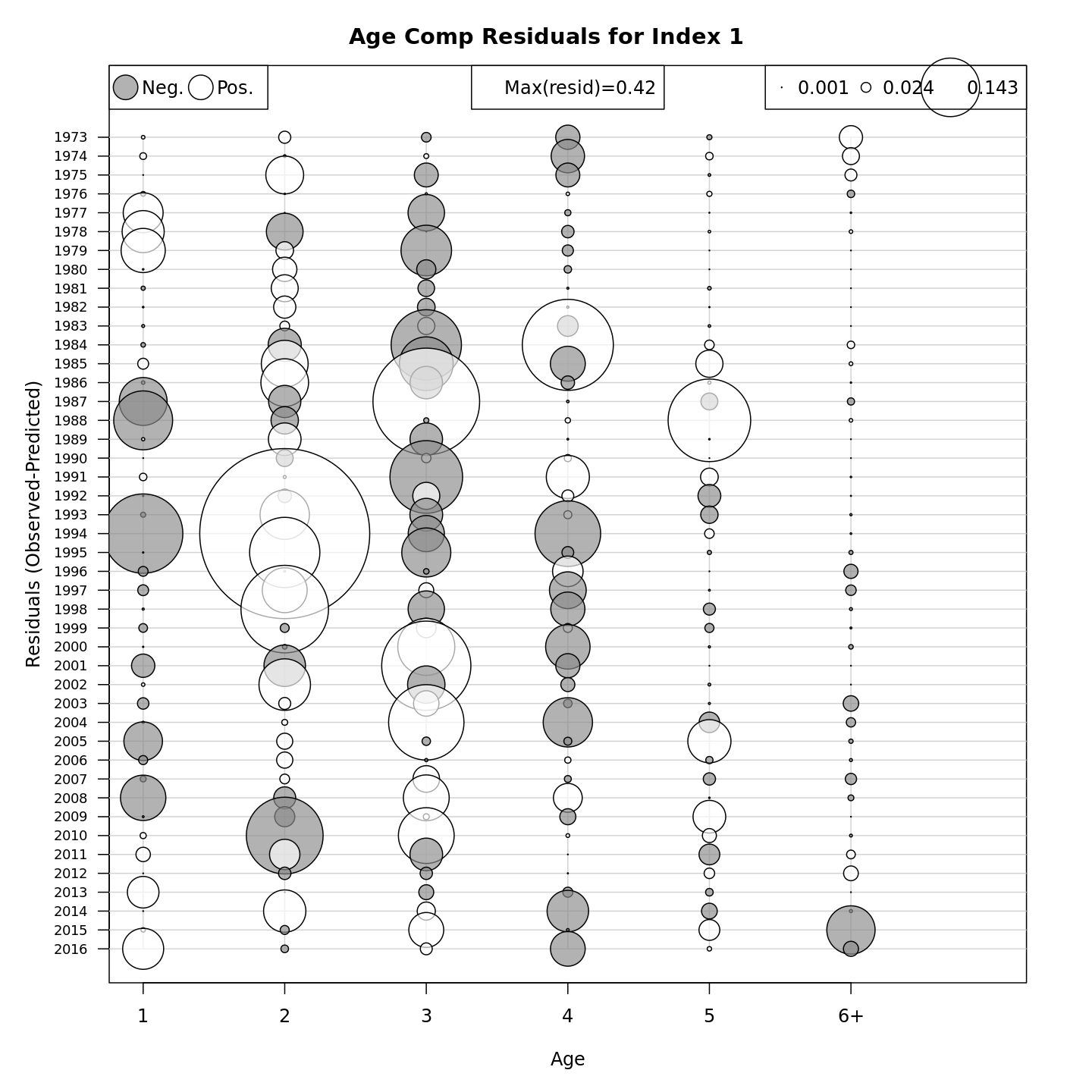
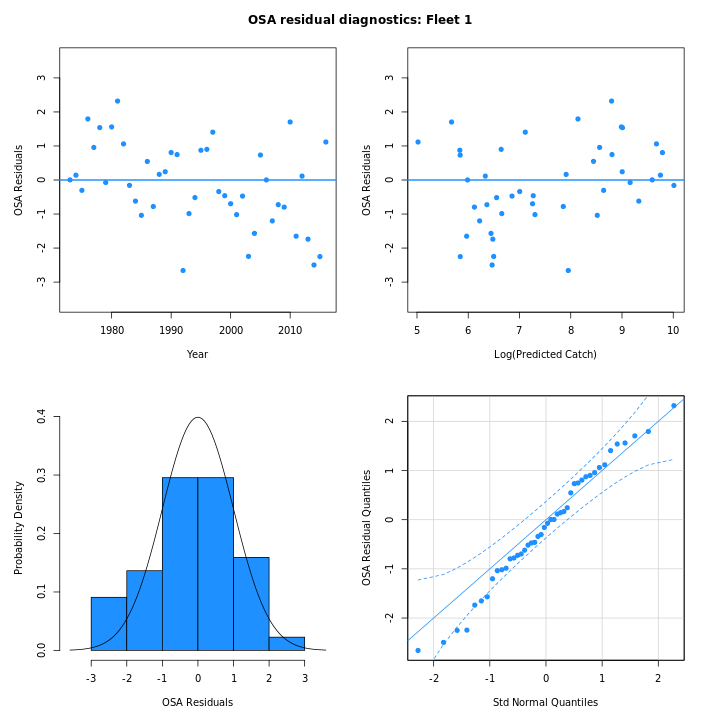
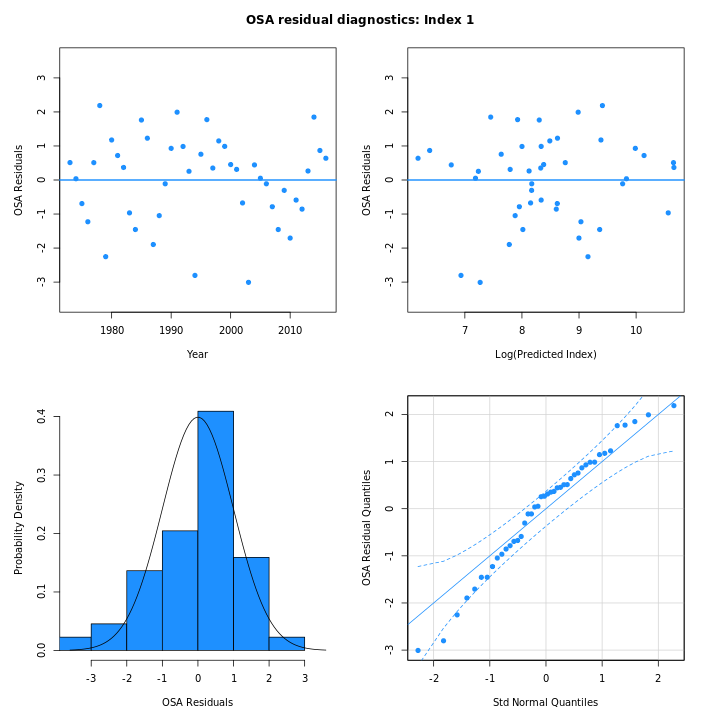
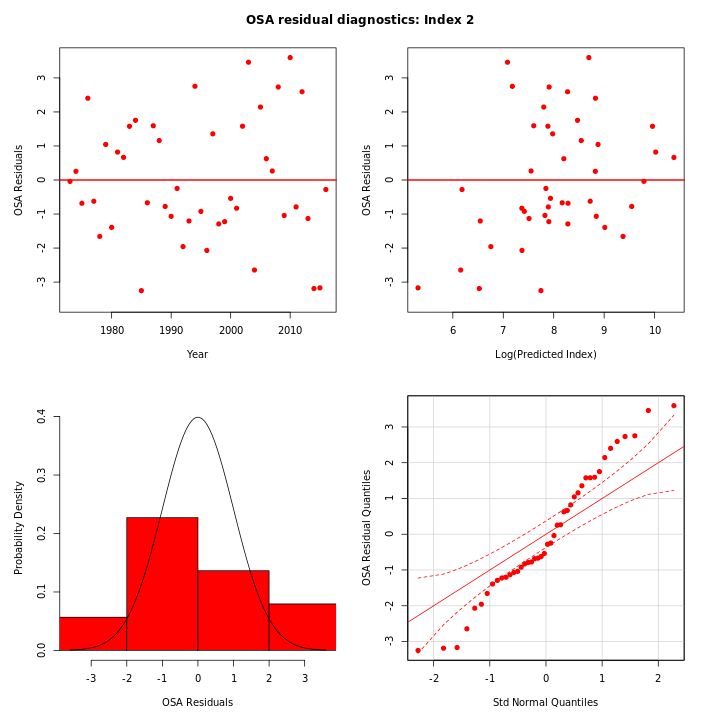
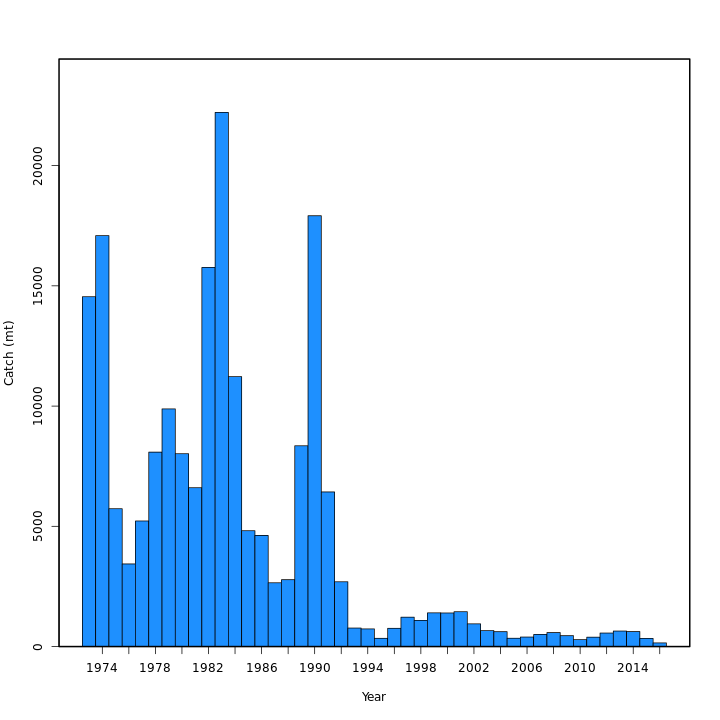
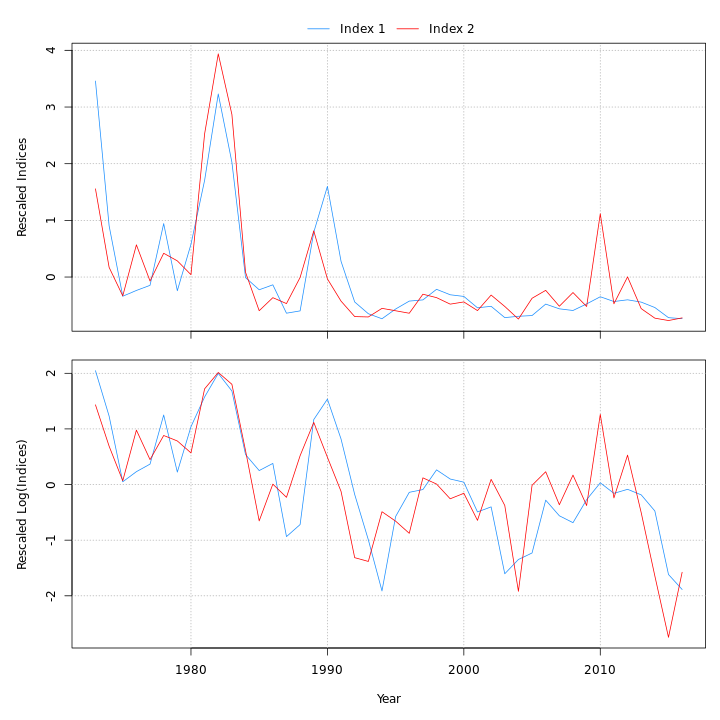
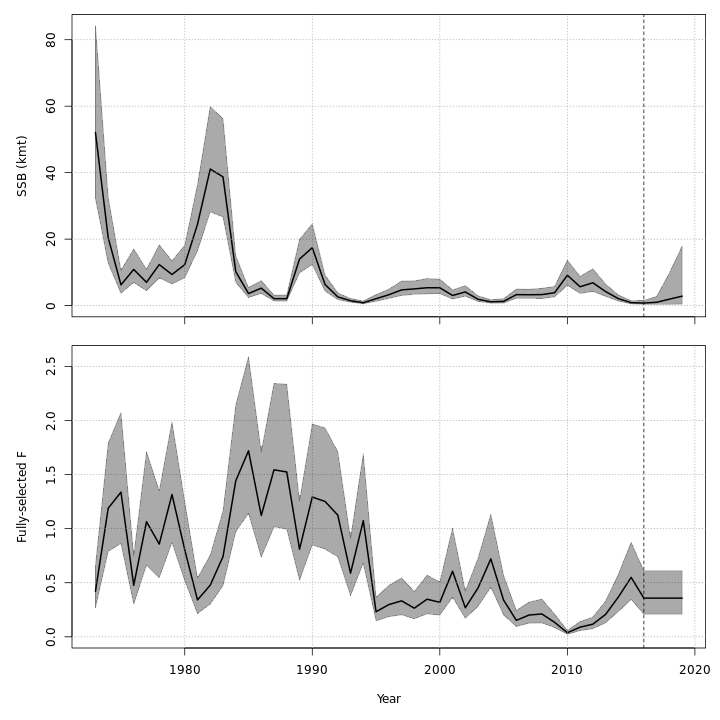
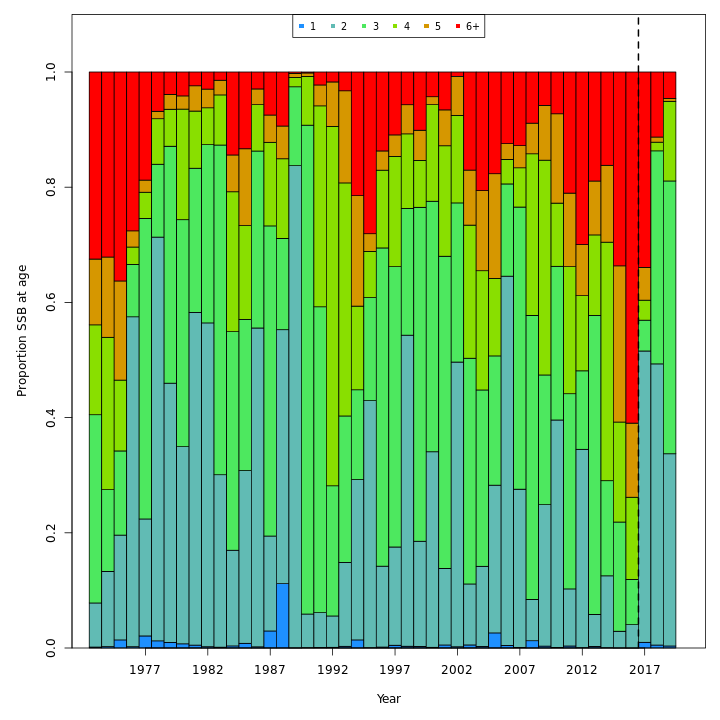
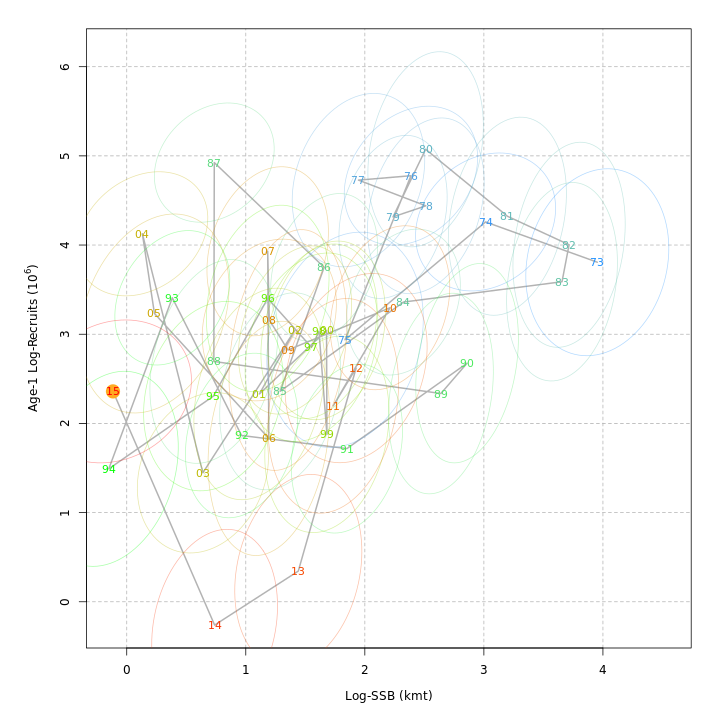
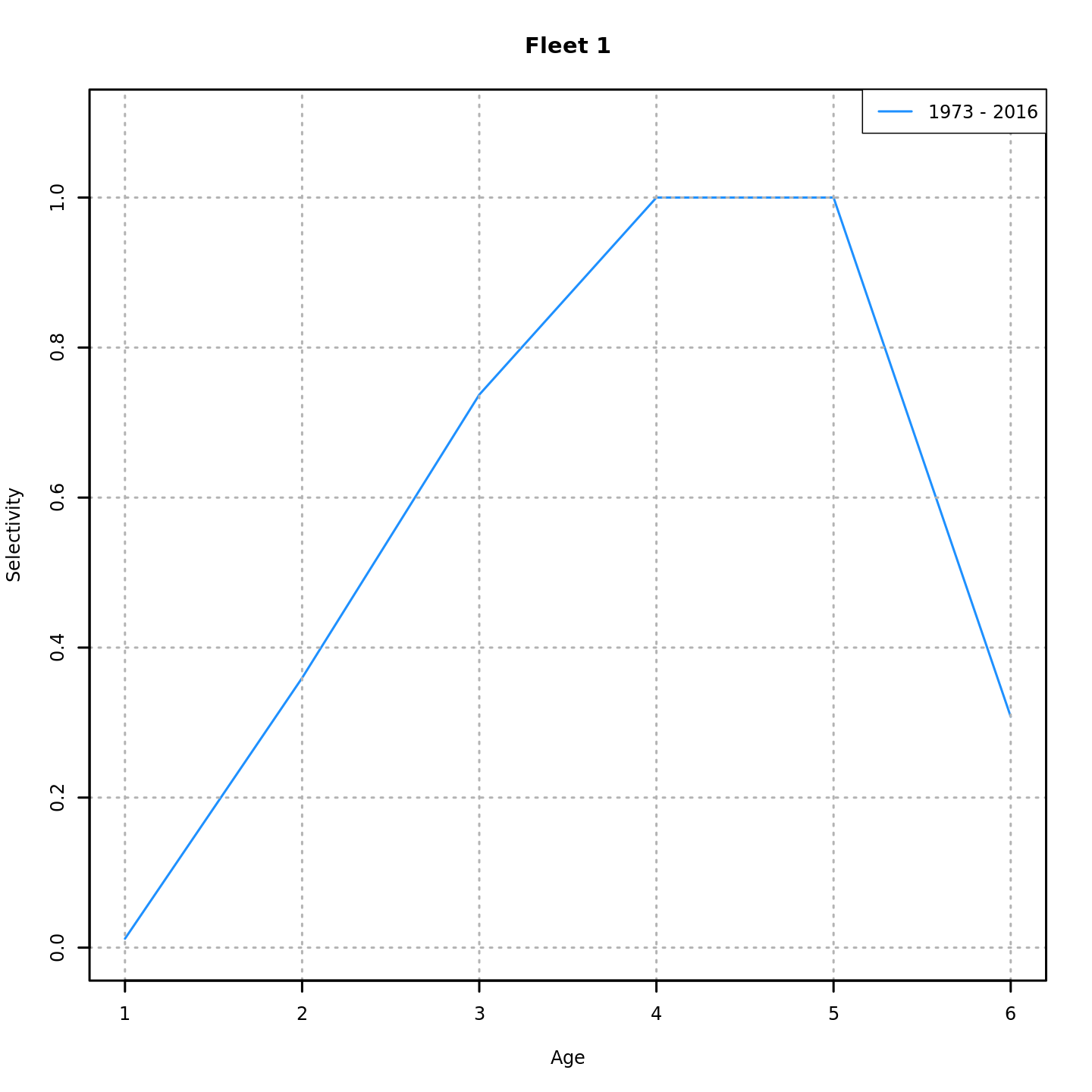
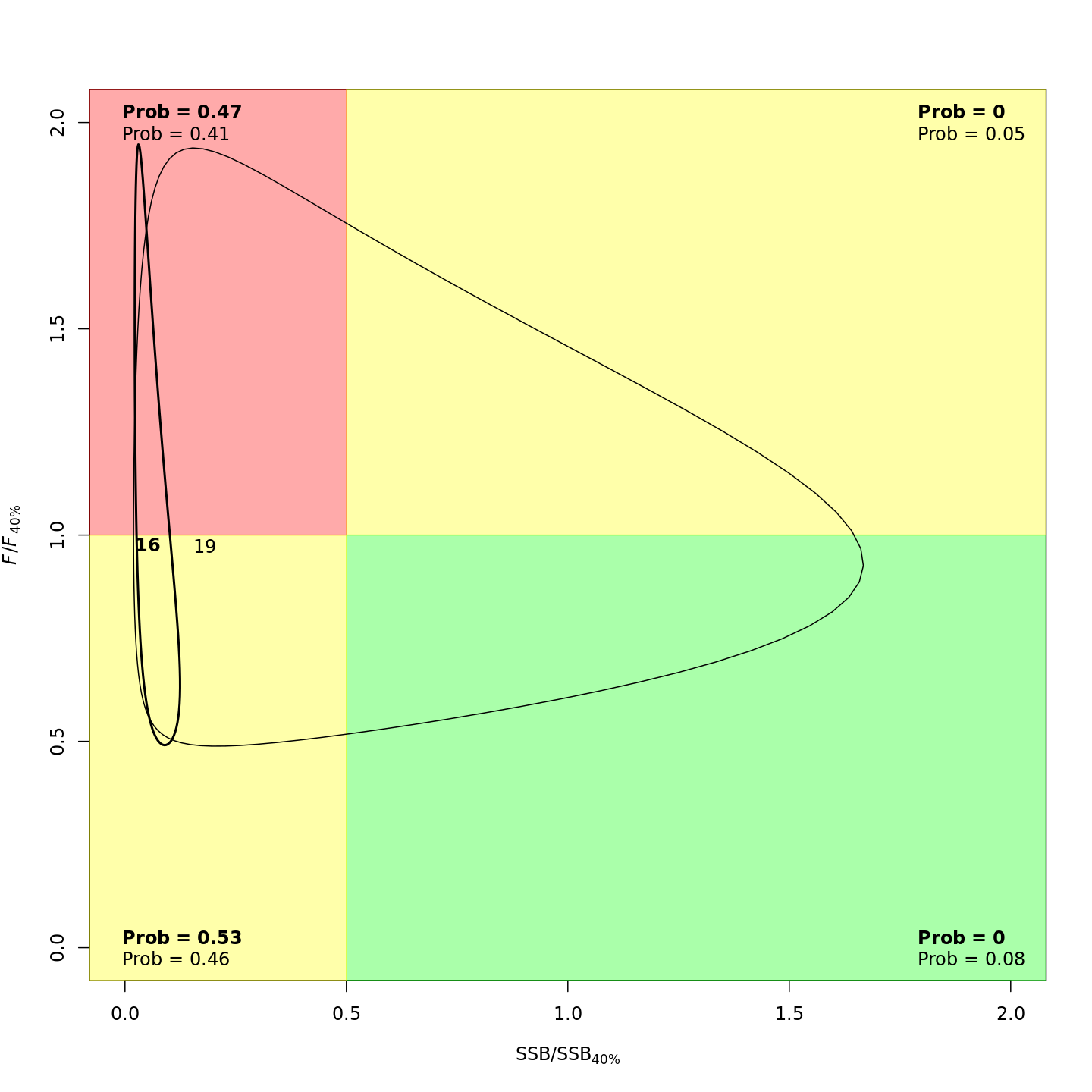
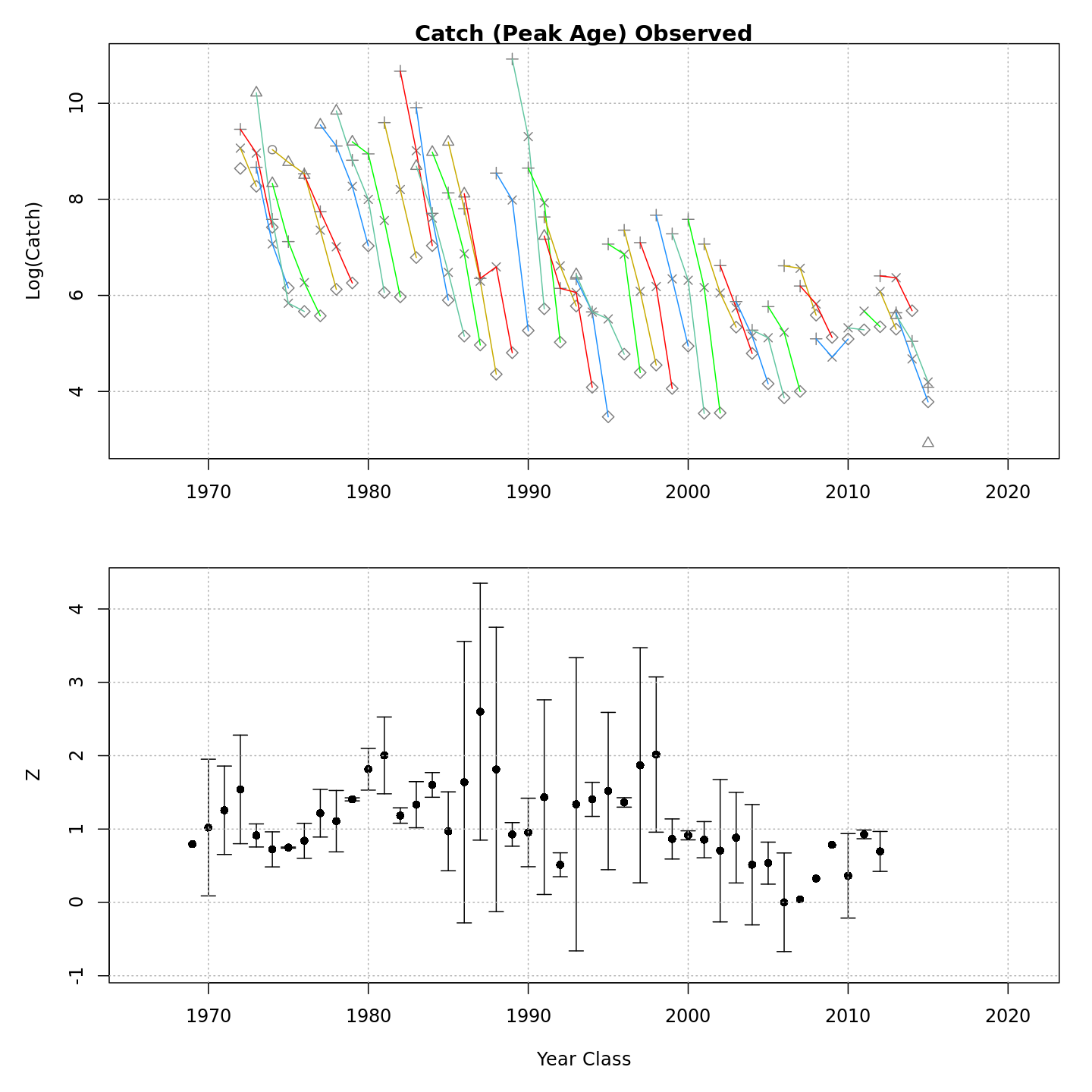
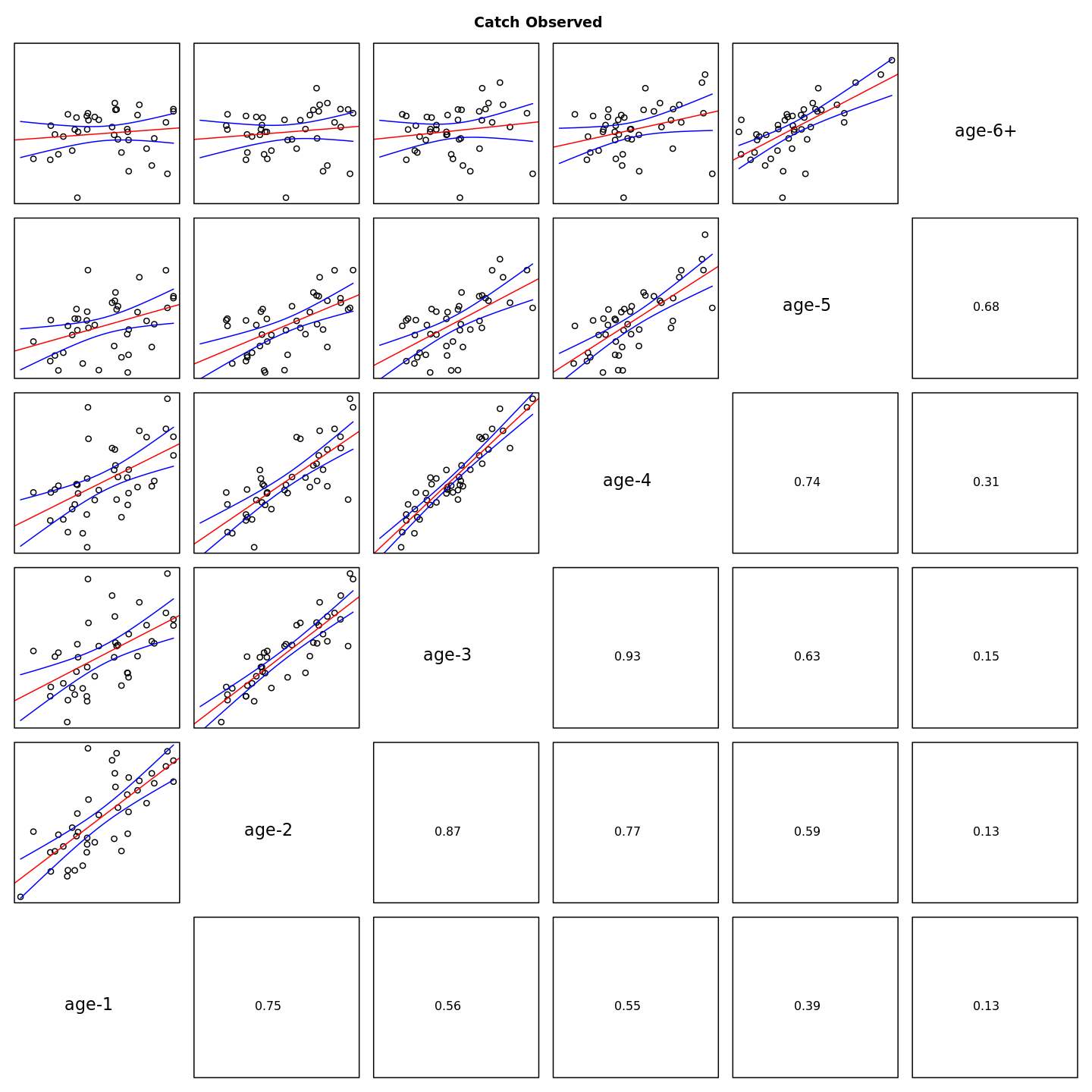
Many plots are generated—here we display some examples: